Scientific Computing
Numerical simulation of real-world phenomena provides fertile ground for building interdisciplinary relationships. The SCI Institute has a long tradition of building these relationships in a win-win fashion – a win for the theoretical and algorithmic development of numerical modeling and simulation techniques and a win for the discipline-specific science of interest. High-order and adaptive methods, uncertainty quantification, complexity analysis, and parallelization are just some of the topics being investigated by SCI faculty. These areas of computing are being applied to a wide variety of engineering applications ranging from fluid mechanics and solid mechanics to bioelectricity.
Valerio Pascucci
Scientific Data Management
Chris Johnson
Problem Solving Environments
Ross Whitaker
GPUs
Chuck Hansen
GPUsFunded Research Projects:
Publications in Scientific Computing:
  Efficient I/O and storage of adaptive-resolution data S. Kumar, J. Edwards, P.-T. Bremer, A. Knoll, C. Christensen, V. Vishwanath, P. Carns, J.A. Schmidt, V. Pascucci. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, IEEE Press, pp. 413--423. 2014. DOI: 10.1109/SC.2014.39 We present an efficient, flexible, adaptive-resolution I/O framework that is suitable for both uniform and Adaptive Mesh Refinement (AMR) simulations. In an AMR setting, current solutions typically represent each resolution level as an independent grid which often results in inefficient storage and performance. Our technique coalesces domain data into a unified, multiresolution representation with fast, spatially aggregated I/O. Furthermore, our framework easily extends to importance-driven storage of uniform grids, for example, by storing regions of interest at full resolution and nonessential regions at lower resolution for visualization or analysis. Our framework, which is an extension of the PIDX framework, achieves state of the art disk usage and I/O performance regardless of resolution of the data, regions of interest, and the number of processes that generated the data. We demonstrate the scalability and efficiency of our framework using the Uintah and S3D large-scale combustion codes on the Mira and Edison supercomputers. |
  Systematic Debugging Methods for Large-Scale HPC Computational Frameworks A. Humphrey, Q. Meng, M. Berzins, D. Caminha B.de Oliveira, Z. Rakamaric, G. Gopalakrishnan. In Computing in Science Engineering, Vol. 16, No. 3, pp. 48--56. May, 2014. ISSN: 1521-9615 DOI: 10.1109/MCSE.2014.11 Parallel computational frameworks for high performance computing (HPC) are central to the advancement of simulation based studies in science and engineering. Unfortunately, finding and fixing bugs in these frameworks can be extremely time consuming. Left unchecked, these bugs can drastically diminish the amount of new science that can be performed. This paper presents our systematic study of the Uintah Computational Framework, and our approaches to debug it more incisively. Our key insight is to leverage the modular structure of Uintah which lends itself to systematic debugging. In particular, we have developed a new approach based on Coalesced Stack Trace Graphs (CSTGs) that summarize the system behavior in terms of key control flows manifested through function invocation chains. We illustrate several scenarios how CSTGs could help efficiently localize bugs, and present a case study of how we found and fixed a real Uintah bug using CSTGs. Keywords: Computational Modeling and Frameworks, Parallel Programming, Reliability, Debugging Aids |
  ASCAC Workforce Subcommittee Letter B. Chapman, H. Calandra, S. Crivelli, J. Dongarra, J. Hittinger, C.R. Johnson, S.A. Lathrop, V. Sarkar, E. Stahlberg, J.S. Vetter, D. Williams. Note: Office of Scientific and Technical Information, DOE ASCAC Committee Report, July, 2014. DOI: 10.2172/1222711 Simulation and computing are essential to much of the research conducted at the DOE national laboratories. Experts in the ASCR-relevant Computing Sciences, which encompass a range of disciplines including Computer Science, Applied Mathematics, Statistics and domain sciences, are an essential element of the workforce in nearly all of the DOE national laboratories. This report seeks to identify the gaps and challenges facing DOE with respect to this workforce. |
  A survey of high level frameworks in block-structured adaptive mesh refinement packages A. Dubey, A. Almgren, John Bell, M. Berzins, S. Brandt, G. Bryan, P. Colella, D. Graves, M. Lijewski, F. Löffler, B. O’Shea, E. Schnetter, B. Van Straalen, K. Weide. In Journal of Parallel and Distributed Computing, 2014. DOI: 10.1016/j.jpdc.2014.07.001 Over the last decade block-structured adaptive mesh refinement (SAMR) has found increasing use in large, publicly available codes and frameworks. SAMR frameworks have evolved along different paths. Some have stayed focused on specific domain areas, others have pursued a more general functionality, providing the building blocks for a larger variety of applications. In this survey paper we examine a representative set of SAMR packages and SAMR-based codes that have been in existence for half a decade or more, have a reasonably sized and active user base outside of their home institutions, and are publicly available. The set consists of a mix of SAMR packages and application codes that cover a broad range of scientific domains. We look at their high-level frameworks, their design trade-offs and their approach to dealing with the advent of radical changes in hardware architecture. The codes included in this survey are BoxLib, Cactus, Chombo, Enzo, FLASH, and Uintah. Keywords: SAMR, BoxLib, Chombo, FLASH, Cactus, Enzo, Uintah |
  Scalable large-scale fluid-structure interaction solvers in the Uintah framework via hybrid task-based parallelism algorithms Q. Meng, M. Berzins. In Concurrency and Computation: Practice and Experience, Vol. 26, No. 7, pp. 1388--1407. May, 2014. DOI: 10.1002/cpe Uintah is a software framework that provides an environment for solving fluid–structure interaction problems on structured adaptive grids for large-scale science and engineering problems involving the solution of partial differential equations. Uintah uses a combination of fluid flow solvers and particle-based methods for solids, together with adaptive meshing and a novel asynchronous task-based approach with fully automated load balancing. When applying Uintah to fluid–structure interaction problems, the combination of adaptive mesh- ing and the movement of structures through space present a formidable challenge in terms of achieving scalability on large-scale parallel computers. The Uintah approach to the growth of the number of core counts per socket together with the prospect of less memory per core is to adopt a model that uses MPI to communicate between nodes and a shared memory model on-node so as to achieve scalability on large-scale systems. For this approach to be successful, it is necessary to design data structures that large numbers of cores can simultaneously access without contention. This scalability challenge is addressed here for Uintah, by the development of new hybrid runtime and scheduling algorithms combined with novel lock-free data structures, making it possible for Uintah to achieve excellent scalability for a challenging fluid–structure problem with mesh refinement on as many as 260K cores. Keywords: MPI, threads, Uintah, many core, lock free, fluid-structure interaction, c-safe |
 International Journal for Uncertainty Quantification, Subtitled “Special Issue on Working with Uncertainty: Representation, Quantification, Propagation, Visualization, and Communication of Uncertainty,” C.R. Johnson, A. Pang (Eds.). In Int. J. Uncertainty Quantification, Vol. 3, No. 3, Begell House, Inc., 2013. ISSN: 2152-5080 DOI: 10.1615/Int.J.UncertaintyQuantification.v3.i3 |
 International Journal for Uncertainty Quantification, Subtitled “Special Issue on Working with Uncertainty: Representation, Quantification, Propagation, Visualization, and Communication of Uncertainty,” C.R. Johnson, A. Pang (Eds.). In Int. J. Uncertainty Quantification, Vol. 3, No. 2, Begell House, Inc., pp. vii--viii. 2013. ISSN: 2152-5080 DOI: 10.1615/Int.J.UncertaintyQuantification.v3.i2 |
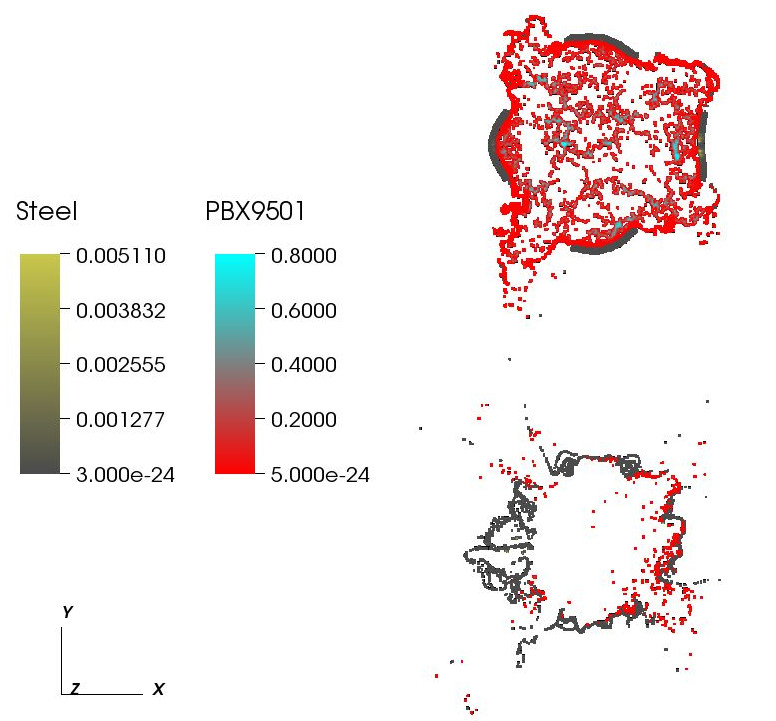
  The influence of an applied heat flux on the violence of reaction of an explosive device M. Hall, J.C. Beckvermit, C.A. Wight, T. Harman, M. Berzins. In Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery, San Diego, California, XSEDE '13, pp. 11:1--11:8. 2013. ISBN: 978-1-4503-2170-9 DOI: 10.1145/2484762.2484786 It is well known that the violence of slow cook-off explosions can greatly exceed the comparatively mild case burst events typically observed for rapid heating. However, there have been few studies that examine the reaction violence as a function of applied heat flux that explore the dependence on heating geometry and device size. Here we report progress on a study using the Uintah Computation Framework, a high-performance computer model capable of modeling deflagration, material damage, deflagration to detonation transition and detonation for PBX9501 and similar explosives. Our results suggests the existence of a sharp threshold for increased reaction violence with decreasing heat flux. The critical heat flux was seen to increase with increasing device size and decrease with the heating of multiple surfaces, suggesting that the temperature gradient in the heated energetic material plays an important role the violence of reactions. Keywords: DDT, cook-off, deflagration, detonation, violence of reaction, c-safe |