Scientific Computing
Numerical simulation of real-world phenomena provides fertile ground for building interdisciplinary relationships. The SCI Institute has a long tradition of building these relationships in a win-win fashion – a win for the theoretical and algorithmic development of numerical modeling and simulation techniques and a win for the discipline-specific science of interest. High-order and adaptive methods, uncertainty quantification, complexity analysis, and parallelization are just some of the topics being investigated by SCI faculty. These areas of computing are being applied to a wide variety of engineering applications ranging from fluid mechanics and solid mechanics to bioelectricity.
Valerio Pascucci
Scientific Data Management
Chris Johnson
Problem Solving Environments
Ross Whitaker
GPUs
Chuck Hansen
GPUsFunded Research Projects:
Publications in Scientific Computing:
 Specimen-specific predictions of contact stress under physiological loading in the human hip: validation and sensitivity studies C.R. Henak, A.K. Kapron, B.J. Ellis, S.A. Maas, A.E. Anderson, J.A. Weiss. In Biomechanics and Modeling in Mechanobiology, pp. 1-14. 2013. DOI: 10.1007/s10237-013-0504-1 Hip osteoarthritis may be initiated and advanced by abnormal cartilage contact mechanics, and finite element (FE) modeling provides an approach with the potential to allow the study of this process. Previous FE models of the human hip have been limited by single specimen validation and the use of quasi-linear or linear elastic constitutive models of articular cartilage. The effects of the latter assumptions on model predictions are unknown, partially because data for the instantaneous behavior of healthy human hip cartilage are unavailable. The aims of this study were to develop and validate a series of specimen-specific FE models, to characterize the regional instantaneous response of healthy human hip cartilage in compression, and to assess the effects of material nonlinearity, inhomogeneity and specimen-specific material coefficients on FE predictions of cartilage contact stress and contact area. Five cadaveric specimens underwent experimental loading, cartilage material characterization and specimen-specific FE modeling. Cartilage in the FE models was represented by average neo-Hookean, average Veronda Westmann and specimen- and region-specific Veronda Westmann hyperelastic constitutive models. Experimental measurements and FE predictions compared well for all three cartilage representations, which was reflected in average RMS errors in contact stress of less than 25 %. The instantaneous material behavior of healthy human hip cartilage varied spatially, with stiffer acetabular cartilage than femoral cartilage and stiffer cartilage in lateral regions than in medial regions. The Veronda Westmann constitutive model with average material coefficients accurately predicted peak contact stress, average contact stress, contact area and contact patterns. The use of subject- and region-specific material coefficients did not increase the accuracy of FE model predictions. The neo-Hookean constitutive model underpredicted peak contact stress in areas of high stress. The results of this study support the use of average cartilage material coefficients in predictions of cartilage contact stress and contact area in the normal hip. The regional characterization of cartilage material behavior provides the necessary inputs for future computational studies, to investigate other mechanical parameters that may be correlated with OA and cartilage damage in the human hip. In the future, the results of this study can be applied to subject-specific models to better understand how abnormal hip contact stress and contact area contribute to OA. |
  Relationship of the intercondylar roof and the tibial footprint of the ACL: implications for ACL reconstruction P.T. Scheffel, H.B. Henninger, R.T. Burks. In American Journal of Sports Medicine, Vol. 41, No. 2, pp. 396--401. 2013. DOI: 10.1177/0363546512467955 Background: Debate exists on the proper relation of the anterior cruciate ligament (ACL) footprint with the intercondylar notch in anatomic ACL reconstructions. Patient-specific graft placement based on the inclination of the intercondylar roof has been proposed. The relationship between the intercondylar roof and native ACL footprint on the tibia has not previously been quantified. Hypothesis: No statistical relationship exists between the intercondylar roof angle and the location of the native footprint of the ACL on the tibia. Study Design: Case series; Level of evidence, 4. Methods: Knees from 138 patients with both lateral radiographs and MRI, without a history of ligamentous injury or fracture, were reviewed to measure the intercondylar roof angle of the femur. Roof angles were measured on lateral radiographs. The MRI data of the same knees were analyzed to measure the position of the central tibial footprint of the ACL (cACL). The roof angle and tibial footprint were evaluated to determine if statistical relationships existed. Results: Patients had a mean ± SD age of 40 ± 16 years. Average roof angle was 34.7° ± 5.2° (range, 23°-48°; 95% CI, 33.9°-35.5°), and it differed by sex but not by side (right/left). The cACL was 44.1% ± 3.4% (range, 36.1%-51.9%; 95% CI, 43.2%-45.0%) of the anteroposterior length of the tibia. There was only a weak correlation between the intercondylar roof angle and the cACL (R = 0.106). No significant differences arose between subpopulations of sex or side. Conclusion: The tibial footprint of the ACL is located in a position on the tibia that is consistent and does not vary according to intercondylar roof angle. The cACL is consistently located between 43.2% and 45.0% of the anteroposterior length of the tibia. Intercondylar roof–based guidance may not predictably place a tibial tunnel in the native ACL footprint. Use of a generic ACL footprint to place a tibial tunnel during ACL reconstruction may be reliable in up to 95% of patients. |
 Evaluation of a post-processing approach for multiscale analysis of biphasic mechanics of chondrocytes S.C. Sibole, S.A. Maas, J.P. Halloran, J.A. Weiss, A. Erdemir. In Computer Methods in Biomechanical and Biomedical Engineering, Vol. 16, No. 10, pp. 1112--1126. 2013. DOI: 10.1080/10255842.2013.809711 PubMed ID: 23809004 Understanding the mechanical behaviour of chondrocytes as a result of cartilage tissue mechanics has significant implications for both evaluation of mechanobiological function and to elaborate on damage mechanisms. A common procedure for prediction of chondrocyte mechanics (and of cell mechanics in general) relies on a computational post-processing approach where tissue-level deformations drive cell-level models. Potential loss of information in this numerical coupling approach may cause erroneous cellular-scale results, particularly during multiphysics analysis of cartilage. The goal of this study was to evaluate the capacity of first- and second-order data passing to predict chondrocyte mechanics by analysing cartilage deformations obtained for varying complexity of loading scenarios. A tissue-scale model with a sub-region incorporating representation of chondron size and distribution served as control. The post-processing approach first required solution of a homogeneous tissue-level model, results of which were used to drive a separate cell-level model (same characteristics as the sub-region of control model). The first-order data passing appeared to be adequate for simplified loading of the cartilage and for a subset of cell deformation metrics, for example, change in aspect ratio. The second-order data passing scheme was more accurate, particularly when asymmetric permeability of the tissue boundaries was considered. Yet, the method exhibited limitations for predictions of instantaneous metrics related to the fluid phase, for example, mass exchange rate. Nonetheless, employing higher order data exchange schemes may be necessary to understand the biphasic mechanics of cells under lifelike tissue loading states for the whole time history of the simulation. |
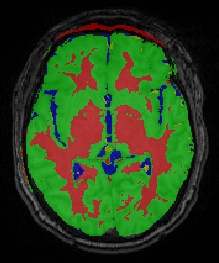
  Evaluation of Current Algorithms for Segmentation of Scar Tissue from Late Gadolinium Enhancement Cardiovascular Magnetic Resonance of the Left Atrium: An Open-Access Grand Challenge R. Karim, R.J. Housden, M. Balasubramaniam, Z. Chen, D. Perry, A. Uddin, Y. Al-Beyatti, E. Palkhi, P. Acheampong, S. Obom, A. Hennemuth, Y. Lu, W. Bai, W. Shi, Y. Gao, H.-O. Peitgen, P. Radau, R. Razavi, A. Tannenbaum, D. Rueckert, J. Cates, T. Schaeffter, D. Peters, R.S. MacLeod, K. Rhode. In Journal of Cardiovascular Magnetic Resonance, Vol. 15, No. 105, 2013. DOI: 10.1186/1532-429X-15-105 Background: Late Gadolinium enhancement (LGE) cardiovascular magnetic resonance (CMR) imaging can be used to visualise regions of fibrosis and scarring in the left atrium (LA) myocardium. This can be important for treatment stratification of patients with atrial fibrillation (AF) and for assessment of treatment after radio frequency catheter ablation (RFCA). In this paper we present a standardised evaluation benchmarking framework for algorithms segmenting fibrosis and scar from LGE CMR images. The algorithms reported are the response to an open challenge that was put to the medical imaging community through an ISBI (IEEE International Symposium on Biomedical Imaging) workshop. Methods: The image database consisted of 60 multicenter, multivendor LGE CMR image datasets from patients with AF, with 30 images taken before and 30 after RFCA for the treatment of AF. A reference standard for scar and fibrosis was established by merging manual segmentations from three observers. Furthermore, scar was also quantified using 2, 3 and 4 standard deviations (SD) and full-width-at-half-maximum (FWHM) methods. Seven institutions responded to the challenge: Imperial College (IC), Mevis Fraunhofer (MV), Sunnybrook Health Sciences (SY), Harvard/Boston University (HB), Yale School of Medicine (YL), King’s College London (KCL) and Utah CARMA (UTA, UTB). There were 8 different algorithms evaluated in this study. Results: Some algorithms were able to perform significantly better than SD and FWHM methods in both pre- and post-ablation imaging. Segmentation in pre-ablation images was challenging and good correlation with the reference standard was found in post-ablation images. Overlap scores (out of 100) with the reference standard were as follows: Pre: IC = 37, MV = 22, SY = 17, YL = 48, KCL = 30, UTA = 42, UTB = 45; Post: IC = 76, MV = 85, SY = 73, HB = 76, YL = 84, KCL = 78, UTA = 78, UTB = 72. Conclusions: The study concludes that currently no algorithm is deemed clearly better than others. There is scope for further algorithmic developments in LA fibrosis and scar quantification from LGE CMR images. Benchmarking of future scar segmentation algorithms is thus important. The proposed benchmarking framework is made available as open-source and new participants can evaluate their algorithms via a web-based interface. Keywords: Late gadolinium enhancement, Cardiovascular magnetic resonance, Atrial fibrillation, Segmentation, Algorithm benchmarking |
  Investigating Applications Portability with the Uintah DAG-based Runtime System on PetaScale Supercomputers Q. Meng, A. Humphrey, J. Schmidt, M. Berzins. In Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis, pp. 96:1--96:12. 2013. ISBN: 978-1-4503-2378-9 DOI: 10.1145/2503210.2503250 Present trends in high performance computing present formidable challenges for applications code using multicore nodes possibly with accelerators and/or co-processors and reduced memory while still attaining scalability. Software frameworks that execute machine-independent applications code using a runtime system that shields users from architectural complexities offer a possible solution. The Uintah framework for example, solves a broad class of large-scale problems on structured adaptive grids using fluid-flow solvers coupled with particle-based solids methods. Uintah executes directed acyclic graphs of computational tasks with a scalable asynchronous and dynamic runtime system for CPU cores and/or accelerators/co-processors on a node. Uintah's clear separation between application and runtime code has led to scalability increases of 1000x without significant changes to application code. This methodology is tested on three leading Top500 machines; OLCF Titan, TACC Stampede and ALCF Mira using three diverse and challenging applications problems. This investigation of scalability with regard to the different processors and communications performance leads to the overall conclusion that the adaptive DAG-based approach provides a very powerful abstraction for solving challenging multi-scale multi-physics engineering problems on some of the largest and most powerful computers available today. Keywords: Blue Gene/Q, GPU, Xeon Phi, adaptive, application, co-processor, heterogeneous systems, hybrid parallelism, parallel, scalability, software, uintah, NETL |
  Preliminary Experiences with the Uintah Framework on Intel Xeon Phi and Stampede Q. Meng, A. Humphrey, J. Schmidt, M. Berzins. In Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery (XSEDE 2013), San Diego, California, pp. 48:1--48:8. 2013. DOI: 10.1145/2484762.2484779 In this work, we describe our preliminary experiences on the Stampede system in the context of the Uintah Computational Framework. Uintah was developed to provide an environment for solving a broad class of fluid-structure interaction problems on structured adaptive grids. Uintah uses a combination of fluid-flow solvers and particle-based methods, together with a novel asynchronous task-based approach and fully automated load balancing. While we have designed scalable Uintah runtime systems for large CPU core counts, the emergence of heterogeneous systems presents considerable challenges in terms of effectively utilizing additional on-node accelerators and co-processors, deep memory hierarchies, as well as managing multiple levels of parallelism. Our recent work has addressed the emergence of heterogeneous CPU/GPU systems with the design of a Unified heterogeneous runtime system, enabling Uintah to fully exploit these architectures with support for asynchronous, out-of-order scheduling of both CPU and GPU computational tasks. Using this design, Uintah has run at full scale on the Keeneland System and TitanDev. With the release of the Intel Xeon Phi co-processor and the recent availability of the Stampede system, we show that Uintah may be modified to utilize such a co-processor based system. We also explore the different usage models provided by the Xeon Phi with the aim of understanding portability of a general purpose framework like Uintah to this architecture. These usage models range from the pragma based offload model to the more complex symmetric model, utilizing all co-processor and host CPU cores simultaneously. We provide preliminary results of the various usage models for a challenging adaptive mesh refinement problem, as well as a detailed account of our experience adapting Uintah to run on the Stampede system. Our conclusion is that while the Stampede system is easy to use, obtaining high performance from the Xeon Phi co-processors requires a substantial but different investment to that needed for GPU-based systems. Keywords: MIC, Xeon Phi, adaptive, co-processor, heterogeneous systems, hybrid parallelism, parallel, scalability, stampede, uintah, c-safe |
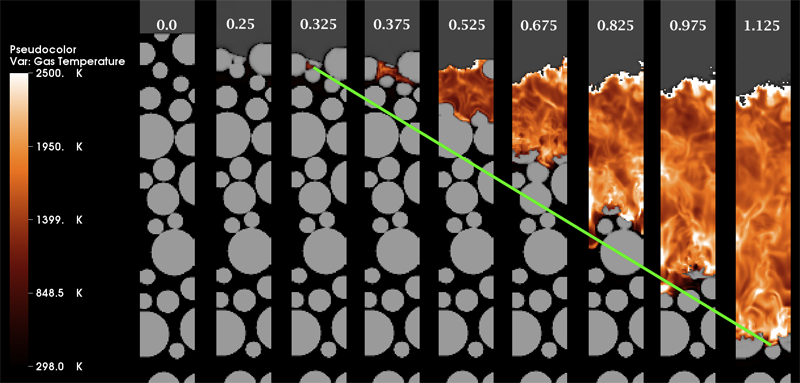
  Multiscale Modeling of Accidental Explosions and Detonations J. Beckvermit, J. Peterson, T. Harman, S. Bardenhagen, C. Wight, Q. Meng, M. Berzins. In Computing in Science and Engineering, Vol. 15, No. 4, pp. 76--86. 2013. DOI: 10.1109/MCSE.2013.89 Accidental explosions are exceptionally dangerous and costly, both in lives and money. Regarding world-wide conflict with small arms and light weapons, the Small Arms Survey has recorded over 297 accidental explosions in munitions depots across the world that have resulted in thousands of deaths and billions of dollars in damage in the past decade alone [45]. As the recent fertilizer plant explosion that killed 15 people in West, Texas demonstrates, accidental explosions are not limited to military operations. Transportation accidents also pose risks, as illustrated by the occasional train derailment/explosion in the nightly news, or the semi-truck explosion detailed in the following section. Unlike other industrial accident scenarios, explosions can easily affect the general public, a dramatic example being the PEPCON disaster in 1988, where windows were shattered, doors blown off their hinges, and flying glass and debris caused injuries up to 10 miles away. While the relative rarity of accidental explosions speaks well of our understanding to date, their violence rightly gives us pause. A better understanding of these materials is clearly still needed, but a significant barrier is the complexity of these materials and the various length scales involved. In typical military applications, explosives are known to be ignited by the coalescence of hot spots which occur on micrometer scales. Whether this reaction remains a deflagration (burning) or builds to a detonation depends both on the stimulus and the boundary conditions or level of confinement. Boundary conditions are typically on the scale of engineered parts, approximately meters. Additional dangers are present at the scale of trucks and factories. The interaction of various entities, such as barrels of fertilizer or crates of detonators, admits the possibility of a sympathetic detonation, i.e. the unintended detonation of one entity by the explosion of another, generally caused by an explosive shock wave or blast fragments. While experimental work has been and will continue to be critical to developing our fundamental understanding of explosive initiation, de agration and detonation, there is no practical way to comprehensively assess safety on the scale of trucks and factories experimentally. The scenarios are too diverse and the costs too great. Numerical simulation provides a complementary tool that, with the steadily increasing computational power of the past decades, makes simulations at this scale begin to look plausible. Simulations at both the micrometer scale, the "mesoscale", and at the scale of engineered parts, the "macro-scale", have been contributing increasingly to our understanding of these materials. Still, simulations on this scale require both massively parallel computational infrastructure and selective sampling of mesoscale response, i.e. advanced computational tools and modeling. The computational framework Uintah [1] has been developed for exactly this purpose. Keywords: uintah, c-safe, accidents, explosions, military computing, risk analysis |
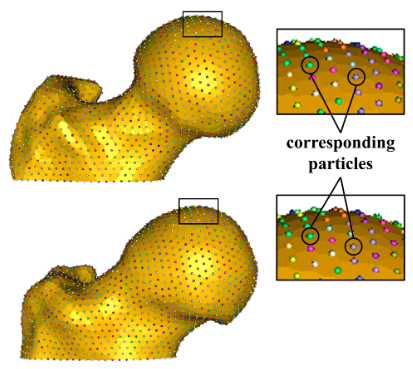
  Statistical Shape Modeling of Cam Femoroacetabular Impingement M.D. Harris, M. Datar, R.T. Whitaker, E.R. Jurrus, C.L. Peters, A.E. Anderson. In Journal of Orthopaedic Research, Vol. 31, No. 10, pp. 1620--1626. 2013. DOI: 10.1002/jor.22389 Statistical shape modeling (SSM) was used to quantify 3D variation and morphologic differences between femurs with and without cam femoroacetabular impingement (FAI). 3D surfaces were generated from CT scans of femurs from 41 controls and 30 cam FAI patients. SSM correspondence particles were optimally positioned on each surface using a gradient descent energy function. Mean shapes for groups were defined. Morphological differences between group mean shapes and between the control mean and individual patients were calculated. Principal component analysis described anatomical variation. Among all femurs, the first six modes (or principal components) captured significant variations, which comprised 84% of cumulative variation. The first two modes, which described trochanteric height and femoral neck width, were significantly different between groups. The mean cam femur shape protruded above the control mean by a maximum of 3.3 mm with sustained protrusions of 2.5–3.0 mm along the anterolateral head-neck junction/distal anterior neck. SSM described variations in femoral morphology that corresponded well with areas prone to damage. Shape variation described by the first two modes may facilitate objective characterization of cam FAI deformities; variation beyond may be inherent population variance. SSM could characterize disease severity and guide surgical resection of bone. |
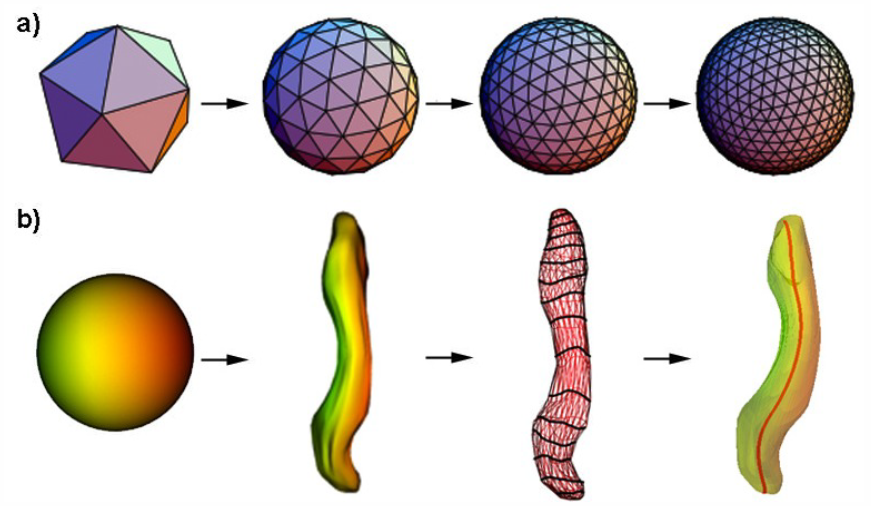
  Lateral ventricle morphology analysis via mean latitude axis B. Paniagua, A. Lyall, J.-B. Berger, C. Vachet, R.M. Hamer, S. Woolson, W. Lin, J. Gilmore, M. Styner. In Proceedings of SPIE 8672, Biomedical Applications in Molecular, Structural, and Functional Imaging, 86720M, 2013. DOI: 10.1117/12.2006846 PubMed ID: 23606800 PubMed Central ID: PMC3630372 Statistical shape analysis has emerged as an insightful method for evaluating brain structures in neuroimaging studies, however most shape frameworks are surface based and thus directly depend on the quality of surface alignment. In contrast, medial descriptions employ thickness information as alignment-independent shape metric. We propose a joint framework that computes local medial thickness information via a mean latitude axis from the well-known spherical harmonic (SPHARM-PDM) shape framework. In this work, we applied SPHARM derived medial representations to the morphological analysis of lateral ventricles in neonates. Mild ventriculomegaly (MVM) subjects are compared to healthy controls to highlight the potential of the methodology. Lateral ventricles were obtained from MRI scans of neonates (9- 144 days of age) from 30 MVM subjects as well as age- and sex-matched normal controls (60 total). SPHARM-PDM shape analysis was extended to compute a mean latitude axis directly from the spherical parameterization. Local thickness and area was straightforwardly determined. MVM and healthy controls were compared using local MANOVA and compared with the traditional SPHARM-PDM analysis. Both surface and mean latitude axis findings differentiate successfully MVM and healthy lateral ventricle morphology. Lateral ventricles in MVM neonates show enlarged shapes in tail and head. Mean latitude axis is able to find significant differences all along the lateral ventricle shape, demonstrating that local thickness analysis provides significant insight over traditional SPHARM-PDM. This study is the first to precisely quantify 3D lateral ventricle morphology in MVM neonates using shape analysis. |
  Modeling 4D changes in pathological anatomy using domain adaptation: analysis of TBI imaging using a tumor database Bo Wang, M. Prastawa, A. Saha, S.P. Awate, A. Irimia, M.C. Chambers, P.M. Vespa, J.D. Van Horn, V. Pascucci, G. Gerig. In Proceedings of the 2013 MICCAI-MBIA Workshop, Lecture Notes in Computer Science (LNCS), Vol. 8159, Note: Awarded Best Paper!, pp. 31--39. 2013. DOI: 10.1007/978-3-319-02126-3_4 Analysis of 4D medical images presenting pathology (i.e., lesions) is signi cantly challenging due to the presence of complex changes over time. Image analysis methods for 4D images with lesions need to account for changes in brain structures due to deformation, as well as the formation and deletion of new structures (e.g., edema, bleeding) due to the physiological processes associated with damage, intervention, and recovery. We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. We demonstrate the effectiveness of our novel approach with the analysis of 4D images of traumatic brain injury (TBI), using a synthetic tumor database as the source domain. |
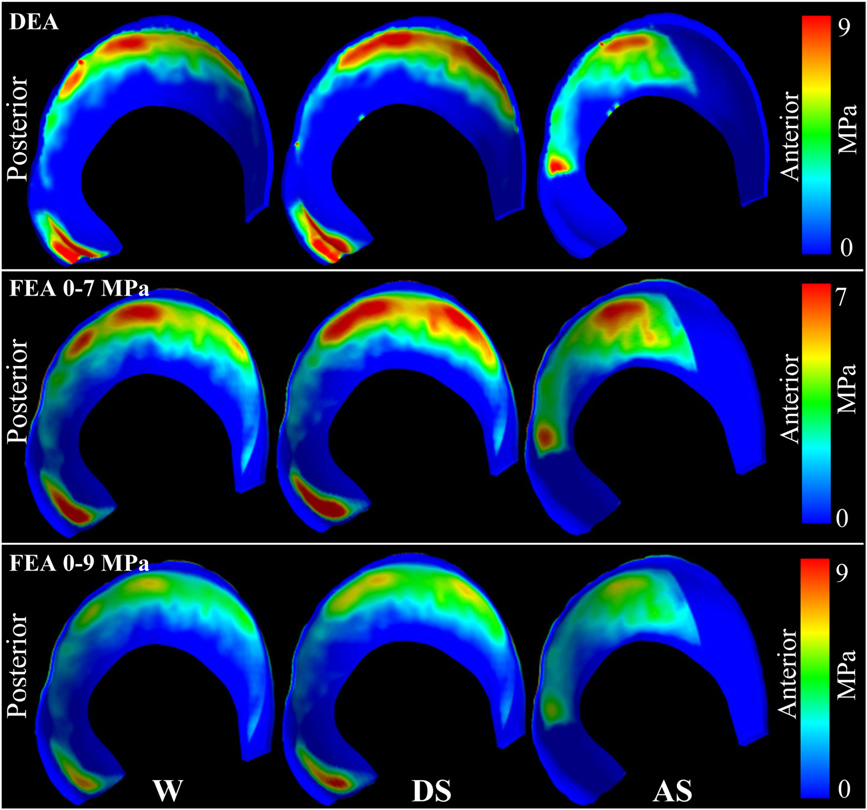
  A new discrete element analysis method for predicting hip joint contact stresses C.L. Abraham, S.A. Maas, J.A. Weiss, B.J. Ellis, C.L. Peters, A.E. Anderson. In Journal of Biomechanics, Vol. 46, No. 6, pp. 1121--1127. 2013. DOI: 10.1016/j.jbiomech.2013.01.012 Quantifying cartilage contact stress is paramount to understanding hip osteoarthritis. Discrete element analysis (DEA) is a computationally efficient method to estimate cartilage contact stresses. Previous applications of DEA have underestimated cartilage stresses and yielded unrealistic contact patterns because they assumed constant cartilage thickness and/or concentric joint geometry. The study objectives were to: (1) develop a DEA model of the hip joint with subject-specific bone and cartilage geometry, (2) validate the DEA model by comparing DEA predictions to those of a validated finite element analysis (FEA) model, and (3) verify both the DEA and FEA models with a linear-elastic boundary value problem. Springs representing cartilage in the DEA model were given lengths equivalent to the sum of acetabular and femoral cartilage thickness and gap distance in the FEA model. Material properties and boundary/loading conditions were equivalent. Walking, descending, and ascending stairs were simulated. Solution times for DEA and FEA models were ∼7 s and ∼65 min, respectively. Irregular, complex contact patterns predicted by DEA were in excellent agreement with FEA. DEA contact areas were 7.5%, 9.7% and 3.7% less than FEA for walking, descending stairs, and ascending stairs, respectively. DEA models predicted higher peak contact stresses (9.8–13.6 MPa) and average contact stresses (3.0–3.7 MPa) than FEA (6.2–9.8 and 2.0–2.5 MPa, respectively). DEA overestimated stresses due to the absence of the Poisson's effect and a direct contact interface between cartilage layers. Nevertheless, DEA predicted realistic contact patterns when subject-specific bone geometry and cartilage thickness were used. This DEA method may have application as an alternative to FEA for pre-operative planning of joint-preserving surgery such as acetabular reorientation during peri-acetabular osteotomy. |
  Three-dimensional Quantification of Femoral Head Shape in Controls and Patients with Cam-type Femoroacetabular Impingement M.D. Harris, S.P. Reese, C.L. Peters, J.A. Weiss, A.E. Anderson. In Annals of Biomedical Engineering, Vol. 41, No. 6, pp. 1162--1171. 2013. DOI: 10.1007/s10439-013-0762-1 An objective measurement technique to quantify 3D femoral head shape was developed and applied to normal subjects and patients with cam-type femoroacetabular impingement (FAI). 3D reconstructions were made from high-resolution CT images of 15 cam and 15 control femurs. Femoral heads were fit to ideal geometries consisting of rotational conchoids and spheres. Geometric similarity between native femoral heads and ideal shapes was quantified. The maximum distance native femoral heads protruded above ideal shapes and the protrusion area were measured. Conchoids provided a significantly better fit to native femoral head geometry than spheres for both groups. Cam-type FAI femurs had significantly greater maximum deviations (4.99 ± 0.39 mm and 4.08 ± 0.37 mm) than controls (2.41 ± 0.31 mm and 1.75 ± 0.30 mm) when fit to spheres or conchoids, respectively. The area of native femoral heads protruding above ideal shapes was significantly larger in controls when a lower threshold of 0.1 mm (for spheres) and 0.01 mm (for conchoids) was used to define a protrusion. The 3D measurement technique described herein could supplement measurements of radiographs in the diagnosis of cam-type FAI. Deviations up to 2.5 mm from ideal shapes can be expected in normal femurs while deviations of 4–5 mm are characteristic of cam-type FAI. |