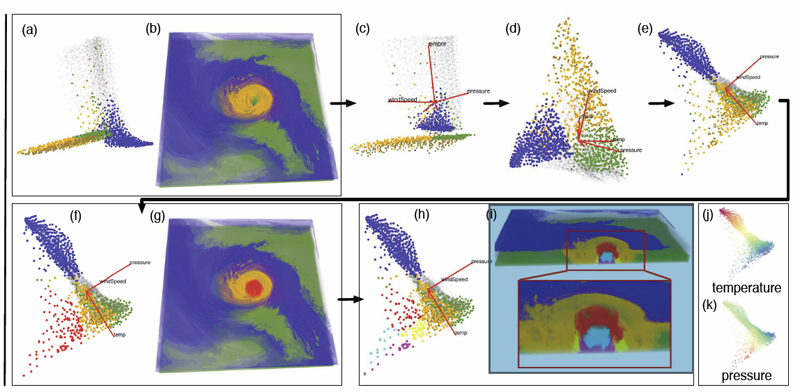
Visualization
Visualization, sometimes referred to as visual data analysis, uses the graphical representation of data as a means of gaining understanding and insight into the data. Visualization research at SCI has focused on applications spanning computational fluid dynamics, medical imaging and analysis, biomedical data analysis, healthcare data analysis, weather data analysis, poetry, network and graph analysis, financial data analysis, etc.Research involves novel algorithm and technique development to building tools and systems that assist in the comprehension of massive amounts of (scientific) data. We also research the process of creating successful visualizations.
We strongly believe in the role of interactivity in visual data analysis. Therefore, much of our research is concerned with creating visualizations that are intuitive to interact with and also render at interactive rates.
Visualization at SCI includes the academic subfields of Scientific Visualization, Information Visualization and Visual Analytics.

Mike Kirby
Uncertainty Visualization
Alex Lex
Information VisualizationCenters and Labs:
- Visualization Design Lab (VDL)
- CEDMAV
- POWDER Display Wall
- Modeling, Display, and Understanding Uncertainty in Simulations for Policy Decision Making
- Topological Data Analysis for Large Network Visualization
Funded Research Projects:
Publications in Visualization:
  Characterizing Cancer Subtypes using Dual Analysis in Caleydo C. Turkay, A. Lex, M. Streit, H. Pfister,, H. Hauser. In IEEE Computer Graphics and Applications, Vol. 34, No. 2, pp. 38--47. March, 2014. ISSN: 0272-1716 DOI: 10.1109/MCG.2014.1 Dual analysis uses statistics to describe both the dimensions and rows of a high-dimensional dataset. Researchers have integrated it into StratomeX, a Caleydo view for cancer subtype analysis. In addition, significant-difference plots show the elements of a candidate subtype that differ significantly from other subtypes, thus letting analysts characterize subtypes. Analysts can also investigate how data samples relate to their assigned subtype and other groups. This approach lets them create well-defined subtypes based on statistical properties. Three case studies demonstrate the approach's utility, showing how it reproduced findings from a published subtype characterization. |
  Mu-8: Visualizing Differences between Proteins and their Families J. Mercer, B. Pandian, A. Lex, N. Bonneel,, H. Pfister. In BMC Proceedings, Vol. 8, No. Suppl 2, pp. S5. Aug, 2014. ISSN: 1753-6561 DOI: 10.1186/1753-6561-8-S2-S5 A complete understanding of the relationship between the amino acid sequence and resulting protein function remains an open problem in the biophysical sciences. Current approaches often rely on diagnosing functionally relevant mutations by determining whether an amino acid frequently occurs at a specific position within the protein family. However, these methods do not account for the biophysical properties and the 3D structure of the protein. We have developed an interactive visualization technique, Mu-8, that provides researchers with a holistic view of the differences of a selected protein with respect to a family of homologous proteins. Mu-8 helps to identify areas of the protein that exhibit: (1) significantly different bio-chemical characteristics, (2) relative conservation in the family, and (3) proximity to other regions that have suspect behavior in the folded protein. |
  RBF Volume Ray Casting on Multicore and Manycore CPUs A. Knoll, I. Wald, P. Navratil, A. Bowen, K. Reda, M. E. Papka, K. Gaither. In Computer Graphics Forum, Vol. 33, No. 3, Edited by H. Carr and P. Rheingans and H. Schumann, Wiley-Blackwell, pp. 71--80. June, 2014. DOI: 10.1111/cgf.12363 Modern supercomputers enable increasingly large N-body simulations using unstructured point data. The structures implied by these points can be reconstructed implicitly. Direct volume rendering of radial basis function (RBF) kernels in domain-space offers flexible classification and robust feature reconstruction, but achieving performant RBF volume rendering remains a challenge for existing methods on both CPUs and accelerators. In this paper, we present a fast CPU method for direct volume rendering of particle data with RBF kernels. We propose a novel two-pass algorithm: first sampling the RBF field using coherent bounding hierarchy traversal, then subsequently integrating samples along ray segments. Our approach performs interactively for a range of data sets from molecular dynamics and astrophysics up to 82 million particles. It does not rely on level of detail or subsampling, and offers better reconstruction quality than structured volume rendering of the same data, exhibiting comparable performance and requiring no additional preprocessing or memory footprint other than the BVH. Lastly, our technique enables multi-field, multi-material classification of particle data, providing better insight and analysis. |
  Overview and State-of-the-Art of Uncertainty Visualization G.P. Bonneau, H.C. Hege, C.R. Johnson, M.M. Oliveira, K. Potter, P. Rheingans, T. Schultz. In Scientific Visualization: Uncertainty, Multifield, Biomedical, and Scalable Visualization, Edited by M. Chen and H. Hagen and C.D. Hansen and C.R. Johnson and A. Kauffman, Springer-Verlag, pp. 3--27. 2014. ISBN: 978-1-4471-6496-8 ISSN: 1612-3786 DOI: 10.1007/978-1-4471-6497-5_1 The goal of visualization is to effectively and accurately communicate data. Visualization research has often overlooked the errors and uncertainty which accompany the scientific process and describe key characteristics used to fully understand the data. The lack of these representations can be attributed, in part, to the inherent difficulty in defining, characterizing, and controlling this uncertainty, and in part, to the difficulty in including additional visual metaphors in a well designed, potent display. However, the exclusion of this information cripples the use of visualization as a decision making tool due to the fact that the display is no longer a true representation of the data. This systematic omission of uncertainty commands fundamental research within the visualization community to address, integrate, and expect uncertainty information. In this chapter, we outline sources and models of uncertainty, give an overview of the state-of-the-art, provide general guidelines, outline small exemplary applications, and finally, discuss open problems in uncertainty visualization. |
  In-situ feature extraction of large scale combustion simulations using segmented merge trees A.G. Landge, V. Pascucci, A. Gyulassy, J.C. Bennett, H. Kolla, J. Chen, P.-T. Bremer. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC 2014), New Orleans, Louisana, IEEE Press, Piscataway, NJ, USA pp. 1020--1031. 2014. ISBN: 978-1-4799-5500-8 DOI: 10.1109/SC.2014.88 The ever increasing amount of data generated by scientific simulations coupled with system I/O constraints are fueling a need for in-situ analysis techniques. Of particular interest are approaches that produce reduced data representations while maintaining the ability to redefine, extract, and study features in a post-process to obtain scientific insights. |
  Efficient I/O and storage of adaptive-resolution data S. Kumar, J. Edwards, P.-T. Bremer, A. Knoll, C. Christensen, V. Vishwanath, P. Carns, J.A. Schmidt, V. Pascucci. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, IEEE Press, pp. 413--423. 2014. DOI: 10.1109/SC.2014.39 We present an efficient, flexible, adaptive-resolution I/O framework that is suitable for both uniform and Adaptive Mesh Refinement (AMR) simulations. In an AMR setting, current solutions typically represent each resolution level as an independent grid which often results in inefficient storage and performance. Our technique coalesces domain data into a unified, multiresolution representation with fast, spatially aggregated I/O. Furthermore, our framework easily extends to importance-driven storage of uniform grids, for example, by storing regions of interest at full resolution and nonessential regions at lower resolution for visualization or analysis. Our framework, which is an extension of the PIDX framework, achieves state of the art disk usage and I/O performance regardless of resolution of the data, regions of interest, and the number of processes that generated the data. We demonstrate the scalability and efficiency of our framework using the Uintah and S3D large-scale combustion codes on the Mira and Edison supercomputers. |
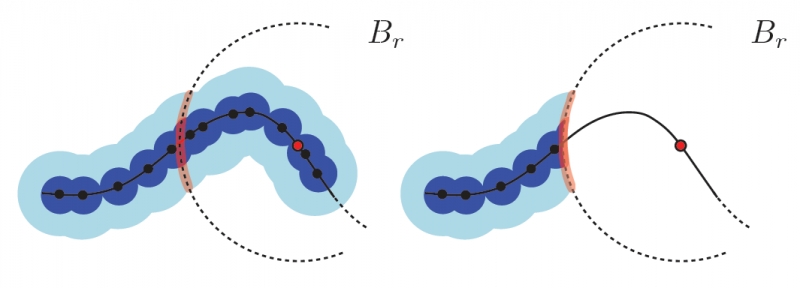
  Robust Detection of Singularities in Vector Fields H. Bhatia, A. Gyulassy, H. Wang, P.-T. Bremer, V. Pascucci . In Topological Methods in Data Analysis and Visualization III, Mathematics and Visualization, Springer International Publishing, pp. 3--18. March, 2014. DOI: 10.1007/978-3-319-04099-8_1 Recent advances in computational science enable the creation of massive datasets of ever increasing resolution and complexity. Dealing effectively with such data requires new analysis techniques that are provably robust and that generate reproducible results on any machine. In this context, combinatorial methods become particularly attractive, as they are not sensitive to numerical instabilities or the details of a particular implementation. We introduce a robust method for detecting singularities in vector fields. We establish, in combinatorial terms, necessary and sufficient conditions for the existence of a critical point in a cell of a simplicial mesh for a large class of interpolation functions. These conditions are entirely local and lead to a provably consistent and practical algorithm to identify cells containing singularities. |
 Scientific Visualization: Uncertainty, Multifield, Biomedical, and Scalable Visualization, C.D. Hansen, M. Chen, C.R. Johnson, A.E. Kaufman, H. Hagen (Eds.). Mathematics and Visualization, Springer, 2014. ISBN: 978-1-4471-6496-8 |
  Information Visualization for Science and Policy: Engaging Users and Avoiding Bias G. McInerny, M. Chen, R. Freeman, D. Gavaghan, M.D. Meyer, F. Rowland, D. Spiegelhalter, M. Steganer, G. Tessarolo, J. Hortal. In Trends in Ecology & Evolution, Vol. 29, No. 3, pp. 148--157. 2014. DOI: 10.1016/j.tree.2014.01.003 Visualisations and graphics are fundamental to studying complex subject matter. However, beyond acknowledging this value, scientists and science-policy programmes rarely consider how visualisations can enable discovery, create engaging and robust reporting, or support online resources. Producing accessible and unbiased visualisations from complicated, uncertain data requires expertise and knowledge from science, policy, computing, and design. However, visualisation is rarely found in our scientific training, organisations, or collaborations. As new policy programmes develop [e.g., the Intergovernmental Platform on Biodiversity and Ecosystem Services (IPBES)], we need information visualisation to permeate increasingly both the work of scientists and science policy. The alternative is increased potential for missed discoveries, miscommunications, and, at worst, creating a bias towards the research that is easiest to display. |