Visualization
Visualization, sometimes referred to as visual data analysis, uses the graphical representation of data as a means of gaining understanding and insight into the data. Visualization research at SCI has focused on applications spanning computational fluid dynamics, medical imaging and analysis, biomedical data analysis, healthcare data analysis, weather data analysis, poetry, network and graph analysis, financial data analysis, etc.Research involves novel algorithm and technique development to building tools and systems that assist in the comprehension of massive amounts of (scientific) data. We also research the process of creating successful visualizations.
We strongly believe in the role of interactivity in visual data analysis. Therefore, much of our research is concerned with creating visualizations that are intuitive to interact with and also render at interactive rates.
Visualization at SCI includes the academic subfields of Scientific Visualization, Information Visualization and Visual Analytics.

Mike Kirby
Uncertainty Visualization
Alex Lex
Information VisualizationCenters and Labs:
- Visualization Design Lab (VDL)
- CEDMAV
- POWDER Display Wall
- Modeling, Display, and Understanding Uncertainty in Simulations for Policy Decision Making
- Topological Data Analysis for Large Network Visualization
Funded Research Projects:
Publications in Visualization:
  Pathfinder: Visual Analysis of Paths in Graphs C. Partl, S. Gratzl, M. Streit, A. Wassermann, H. Pfister, D. Schmalstieg, A. Lex. In Computer Graphics Forum (EuroVis '16), Vol. 35, No. 3, pp. 71-80. jun, 2016. ISSN: 1467-8659 DOI: 10.1111/cgf.12883 The analysis of paths in graphs is highly relevant in many domains. Typically, path-related tasks are performed in node-link layouts. Unfortunately, graph layouts often do not scale to the size of many real world networks. Also, many networks are multivariate, i.e., contain rich attribute sets associated with the nodes and edges. These attributes are often critical in judging paths, but directly visualizing attributes in a graph layout exacerbates the scalability problem. In this paper, we present visual analysis solutions dedicated to path-related tasks in large and highly multivariate graphs. We show that by focusing on paths, we can address the scalability problem of multivariate graph visualization, equipping analysts with a powerful tool to explore large graphs. We introduce Pathfinder, a technique that provides visual methods to query paths, while considering various constraints. The resulting set of paths is visualized in both a ranked list and as a node-link diagram. For the paths in the list, we display rich attribute data associated with nodes and edges, and the node-link diagram provides topological context. The paths can be ranked based on topological properties, such as path length or average node degree, and scores derived from attribute data. Pathfinder is designed to scale to graphs with tens of thousands of nodes and edges by employing strategies such as incremental query results. We demonstrate Pathfinder's fitness for use in scenarios with data from a coauthor network and biological pathways. |
  From Visual Exploration to Storytelling and Back Again Samuel Gratzl, Alexander Lex, Nils Gehlenborg, Nicola Cosgrove, Marc Streit . In Computer Graphics Forum, Vol. 35, No. 3, pp. 491--500. jun, 2016. ISSN: 1467-8659 DOI: 10.1111/cgf.12925 The primary goal of visual data exploration tools is to enable the discovery of new insights. To justify and reproduce insights, the discovery process needs to be documented and communicated. A common approach to documenting and presenting findings is to capture visualizations as images or videos. Images, however, are insufficient for telling the story of a visual discovery, as they lack full provenance information and context. Videos are difficult to produce and edit, particularly due to the non-linear nature of the exploratory process. Most importantly, however, neither approach provides the opportunity to return to any point in the exploration in order to review the state of the visualization in detail or to conduct additional analyses. In this paper we present CLUE (Capture, Label, Understand, Explain), a model that tightly integrates data exploration and presentation of discoveries. Based on provenance data captured during the exploration process, users can extract key steps, add annotations, and author "Vistories", visual stories based on the history of the exploration. These Vistories can be shared for others to view, but also to retrace and extend the original analysis. We discuss how the CLUE approach can be integrated into visualization tools and provide a prototype implementation. Finally, we demonstrate the general applicability of the model in two usage scenarios: a Gapminder-inspired visualization to explore public health data and an example from molecular biology that illustrates how Vistories could be used in scientific journals. |
  TOD-Tree: Task-Overlapped Direct send Tree Image Compositing for Hybrid MPI Parallelism and GPUs A. V. P. Grosset, M. Prasad, C. Christensen, A. Knoll, C. Hansen. In IEEE Transactions on Visualization and Computer Graphics, IEEE, pp. 1--1. 2016. DOI: 10.1109/tvcg.2016.2542069 Modern supercomputers have thousands of nodes, each with CPUs and/or GPUs capable of several teraflops. However, the network connecting these nodes is relatively slow, on the order of gigabits per second. For time-critical workloads such as interactive visualization, the bottleneck is no longer computation but communication. In this paper, we present an image compositing algorithm that works on both CPU-only and GPU-accelerated supercomputers and focuses on communication avoidance and overlapping communication with computation at the expense of evenly balancing the workload. The algorithm has three stages: a parallel direct send stage, followed by a tree compositing stage and a gather stage. We compare our algorithm with radix-k and binary-swap from the IceT library in a hybrid OpenMP/MPI setting on the Stampede and Edison supercomputers, show strong scaling results and explain how we generally achieve better performance than these two algorithms. We developed a GPU-based image compositing algorithm where we use CUDA kernels for computation and GPU Direct RDMA for inter-node GPU communication. We tested the algorithm on the Piz Daint GPU-accelerated supercomputer and show that we achieve performance on par with CPUs. Lastly, we introduce a workflow in which both rendering and compositing are done on the GPU. |
  Kernel Partial Least Squares Regression for Relating Functional Brain Network Topology to Clinical Measures of Behavior E. Wong, S. Palande, Bei Wang, B. Zielinski, J. Anderson, P. T. Fletcher. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), IEEE, April, 2016. DOI: 10.1109/isbi.2016.7493506 In this paper we present a novel method for analyzing the relationship between functional brain networks and behavioral phenotypes. Drawing from topological data analysis, we first extract topological features using persistent homology from functional brain networks that are derived from correlations in resting-state fMRI. Rather than fixing a discrete network topology by thresholding the connectivity matrix, these topological features capture the network organization across all continuous threshold values. We then propose to use a kernel partial least squares (kPLS) regression to statistically quantify the relationship between these topological features and behavior measures. The kPLS also provides an elegant way to combine multiple image features by using linear combinations of multiple kernels. In our experiments we test the ability of our proposed brain network analysis to predict autism severity from rs-fMRI. We show that combining correlations with topological features gives better prediction of autism severity than using correlations alone. |
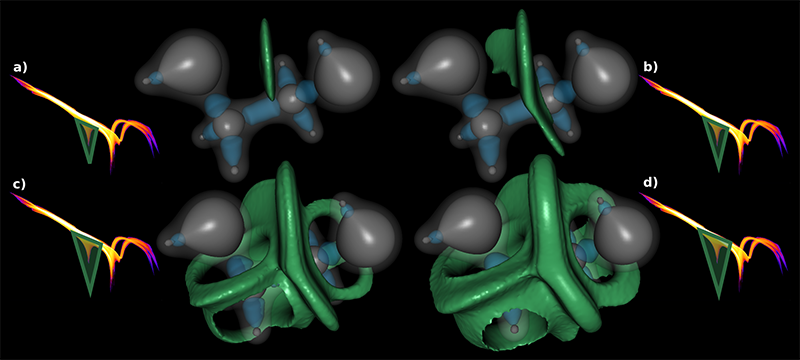
  View-Dependent Streamline Deformation and Exploration X. Tong, J. Edwards, C. Chen, H. Shen, C. R. Johnson, P. Wong. In Transactions on Visualization and Computer Graphics, Vol. 22, No. 7, IEEE, pp. 1788--1801. July, 2016. ISSN: 1077-2626 DOI: 10.1109/tvcg.2015.2502583 Occlusion presents a major challenge in visualizing 3D flow and tensor fields using streamlines. Displaying too many streamlines creates a dense visualization filled with occluded structures, but displaying too few streams risks losing important features. We propose a new streamline exploration approach by visually manipulating the cluttered streamlines by pulling visible layers apart and revealing the hidden structures underneath. This paper presents a customized view-dependent deformation algorithm and an interactive visualization tool to minimize visual clutter in 3D vector and tensor fields. The algorithm is able to maintain the overall integrity of the fields and expose previously hidden structures. Our system supports both mouse and direct-touch interactions to manipulate the viewing perspectives and visualize the streamlines in depth. By using a lens metaphor of different shapes to select the transition zone of the targeted area interactively, the users can move their focus and examine the vector or tensor field freely. Keywords: Context;Deformable models;Lenses;Shape;Streaming media;Three-dimensional displays;Visualization;Flow visualization;deformation;focus+context;occlusion;streamline;white matter tracts |
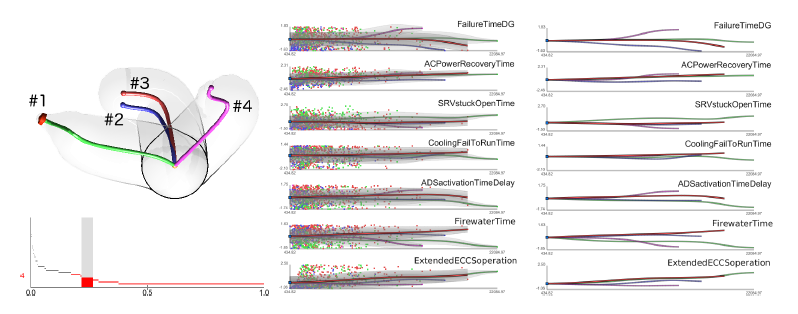
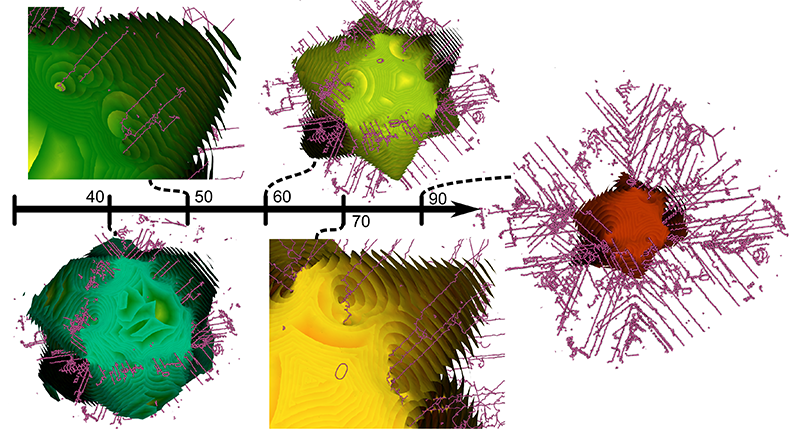
  Analyzing Simulation-Based PRA Data Through Traditional and Topological Clustering: A BWR Station Blackout Case Study D. Maljovec, S. Liu, Bei Wang, V. Pascucci, P. T. Bremer, D. Mandelli, C. Smith.. In Reliability Engineering & System Safety, Vol. 145, Elsevier, pp. 262--276. January, 2016. DOI: 10.1016/j.ress.2015.07.001 Dynamic probabilistic risk assessment (DPRA) methodologies couple system simulator codes (e.g., RELAP, MELCOR) with simulation controller codes (e.g., RAVEN, ADAPT). Whereas system simulator codes model system dynamics deterministically, simulation controller codes introduce both deterministic (e.g., system control logic, operating procedures) and stochastic (e.g., component failures, parameter uncertainties) elements into the simulation. Typically, a DPRA is performed by sampling values of a set of parameters, and simulating the system behavior for that specific set of parameter values. For complex systems, a major challenge in using DPRA methodologies is to analyze the large number of scenarios generated, where clustering techniques are typically employed to better organize and interpret the data. In this paper, we focus on the analysis of two nuclear simulation datasets that are part of the risk-informed safety margin characterization (RISMC) boiling water reactor (BWR) station blackout (SBO) case study. We provide the domain experts a software tool that encodes traditional and topological clustering techniques within an interactive analysis and visualization environment, for understanding the structures of such high-dimensional nuclear simulation datasets. We demonstrate through our case study that both types of clustering techniques complement each other in bringing enhanced structural understanding of the data. |
  muView: A Visual Analysis System for Exploring Uncertainty in Myocardial Ischemia Simulations P. Rosen, B. Burton, K. Potter, C.R. Johnson. In Visualization in Medicine and Life Sciences III, Springer Nature, pp. 49--69. 2016. DOI: 10.1007/978-3-319-24523-2_3 In this paper we describe the Myocardial Uncertainty Viewer (muView or µView) system for exploring data stemming from the simulation of cardiac ischemia. The simulation uses a collection of conductivity values to understand how ischemic regions effect the undamaged anisotropic heart tissue. The data resulting from the simulation is multi-valued and volumetric, and thus, for every data point, we have a collection of samples describing cardiac electrical properties. µView combines a suite of visual analysis methods to explore the area surrounding the ischemic zone and identify how perturbations of variables change the propagation of their effects. In addition to presenting a collection of visualization techniques, which individually highlight different aspects of the data, the coordinated view system forms a cohesive environment for exploring the simulations.We also discuss the findings of our study, which are helping to steer further development of the simulation and strengthening our collaboration with the biomedical engineers attempting to understand the phenomenon. |
  OceanPaths: Visualizing Multivariate Oceanography Data C. Nobre, A. Lex. In Eurographics Conference on Visualization (EuroVis) - Short Papers, Edited by E. Bertini, J. Kennedy, E. Puppo, The Eurographics Association, 2015. DOI: 10.2312/eurovisshort.20151124 Geographical datasets are ubiquitous in oceanography. While map-based visualizations are useful for many different domains, they can suffer from cluttering and overplotting issues when used for multivariate data sets. As a result, spatial data exploration in oceanography has often been restricted to multiple maps showing various depths or time intervals. This lack of interactive exploration often hinders efforts to expose correlations between properties of oceanographic features, specifically currents. OceanPaths provides powerful interaction and exploration methods for spatial, multivariate oceanography datasets to remedy these situations. Fundamentally, our method allows users to define pathways, typically following currents, along which the variation of the high-dimensional data can be plotted efficiently. We present a case study conducted by domain experts to underscore the usefulness of OceanPaths in uncovering trends and correlations in oceanographic data sets. |
  Approximating the Generalized Voronoi Diagram of Closely Spaced Objects J. Edwards, E. Daniel, V. Pascucci, C. Bajaj. In Computer Graphics Forum, Vol. 34, No. 2, Wiley-Blackwell, pp. 299-309. May, 2015. DOI: 10.1111/cgf.12561 Generalized Voronoi Diagrams (GVDs) have far-reaching applications in robotics, visualization, graphics, and simulation. However, while the ordinary Voronoi Diagram has mature and efficient algorithms for its computation, the GVD is difficult to compute in general, and in fact, has only approximation algorithms for anything but the simplest of datasets. Our work is focused on developing algorithms to compute the GVD efficiently and with bounded error on the most difficult of datasets -- those with objects that are extremely close to each other. |
  Paint and Click: Unified Interactions for Image Boundaries B. Summa, A. A. Gooch, G. Scorzelli, V. Pascucci. In Computer Graphics Forum, Vol. 34, No. 2, Wiley-Blackwell, pp. 385--393. May, 2015. DOI: 10.1111/cgf.12568 Image boundaries are a fundamental component of many interactive digital photography techniques, enabling applications such as segmentation, panoramas, and seamless image composition. Interactions for image boundaries often rely on two complementary but separate approaches: editing via painting or clicking constraints. In this work, we provide a novel, unified approach for interactive editing of pairwise image boundaries that combines the ease of painting with the direct control of constraints. Rather than a sequential coupling, this new formulation allows full use of both interactions simultaneously, giving users unprecedented flexibility for fast boundary editing. To enable this new approach, we provide technical advancements. In particular, we detail a reformulation of image boundaries as a problem of finding cycles, expanding and correcting limitations of the previous work. Our new formulation provides boundary solutions for painted regions with performance on par with state-of-the-art specialized, paint-only techniques. In addition, we provide instantaneous exploration of the boundary solution space with user constraints. Finally, we provide examples of common graphics applications impacted by our new approach. |
  An Introduction to Verification of Visualization Techniques T. Etiene, R.M. Kirby, C. Silva. Morgan & Claypool Publishers, 2015. |
  Visualization C.R. Johnson. In Encyclopedia of Applied and Computational Mathematics, Edited by Björn Engquist, Springer, pp. 1537-1546. 2015. ISBN: 978-3-540-70528-4 DOI: 10.1007/978-3-540-70529-1_368 |