Gordon L. Kindlmann, David M. Weinstein, Agatha D. Lee, Arthur W. Toga, Paul M. Thompson
The computation, visualization, and interpretation of brain variability remains a significant challenge in computational neuroanatomy. Current deformable registration methods can generate, for each vertex of a polygonal mesh modeling the cortical surface, a distribution of displacement vectors between the individual models and their average, which can be summarized as a covariance tensor. While analysis of anatomical covariance tensor fields promises insight into the structural components of aging and disease, basic understanding of the tensor field structure is hampered by the lack of effective methods to create informative and interactive visualizations. We describe a novel application of superquadric tensor glyphs to anatomic covariance tensor fields, supplemented by colormaps of important tensor attributes. The resulting visualizations support a more detailed characterization of population variability of brain structure than possible with previous methods, while also suggesting directions for subsequent quantitative analysis.
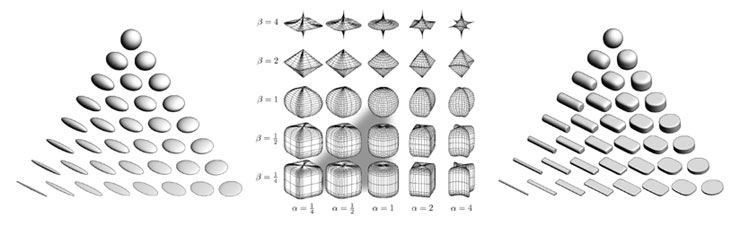 Figure 1: (a) Ellipsoid tensor glyphs, (b) Space of superquadrics, (c) Superquadric tensor glyphs. Ellipsoidal glyphs (a) suffer from visual ambiguity. The range of superquadrics (b) used for tensor glyphs is highlighted with the gray triangle. Superquadric glyphs (c) differentiate shape and convey orientation more clearly than do ellipsoids. Figure 1: (a) Ellipsoid tensor glyphs, (b) Space of superquadrics, (c) Superquadric tensor glyphs. Ellipsoidal glyphs (a) suffer from visual ambiguity. The range of superquadrics (b) used for tensor glyphs is highlighted with the gray triangle. Superquadric glyphs (c) differentiate shape and convey orientation more clearly than do ellipsoids.
|
 Figure 1: (a) Ellipsoid tensor glyphs, (b) Space of superquadrics, (c) Superquadric tensor glyphs. Ellipsoidal glyphs (a) suffer from visual ambiguity. The range of superquadrics (b) used for tensor glyphs is highlighted with the gray triangle. Superquadric glyphs (c) differentiate shape and convey orientation more clearly than do ellipsoids.
Figure 1: (a) Ellipsoid tensor glyphs, (b) Space of superquadrics, (c) Superquadric tensor glyphs. Ellipsoidal glyphs (a) suffer from visual ambiguity. The range of superquadrics (b) used for tensor glyphs is highlighted with the gray triangle. Superquadric glyphs (c) differentiate shape and convey orientation more clearly than do ellipsoids.