 The uses of map3d are varied but include:
The uses of map3d are varied but include:- Visualization and interactive editing of geometric models constructed from line segments, triangles, and tetrahedra. The user can manually remove and edit connectivities between nodes, remove nodes, and manipulate icons that serve as visual landmarks (e.g., blood vessels, cutting planes, and site markers.)
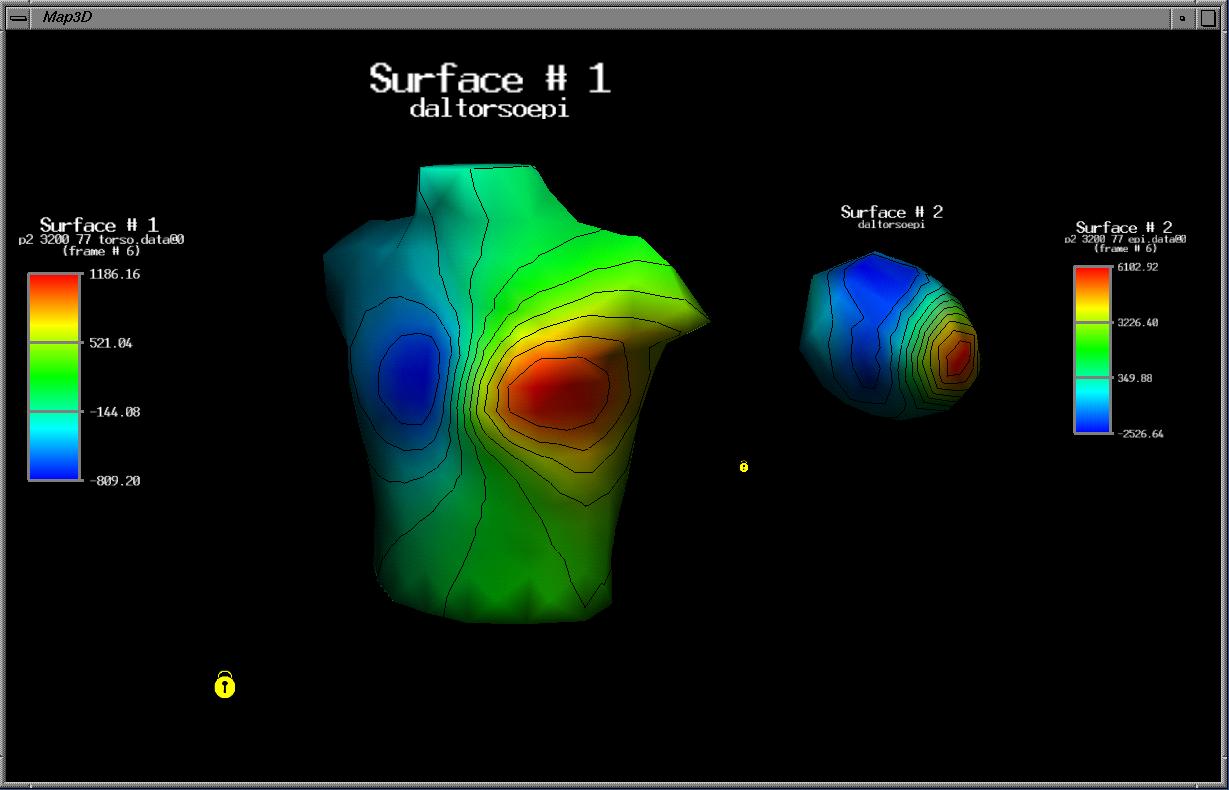
- Visualization of spatially sampled time signals from experiments in electrophysiology; the program offers many scaling options and color maps as well as visualization elements such as surface contours and color-coded surface rendering.
- Simultaneous display of any number of individual time signals.
- Interactive visualization of experimental and simulation results from multiple three-dimensional surfaces displayed either together or in separate windows.
map3d addresses a unique niche in visualization by combining high performance interactive graphics with extensive quantitative data interrogation tools. The user can view three-dimensional time signals in modes that emphasize either the spatial or the temporal aspects of the data and move quickly back and forth between these views.
For more information, check out map3d: Interactive Scientific Visualization for Bioengineering Data.
map3d Acknowledgement
map3d License
Acknowledgement: map3d is an Open Source software project that is principally funded through the SCI Institute's NIH/NIGMS CIBC Center. Please use the following acknowledgment and send us references to any publications, presentations, or successful funding applications that make use of NIH/NIGMS CIBC software or data sets.
"This project was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P41 GM103545 and R24 GM136986.”
• map3d Citation: [bibtex citation] [Endnote citation]
map3d is available for free and open source under the MIT License
The MIT License
Copyright (c) 2007 Scientific Computing and Imaging Institute, University of Utah.
License for the specific language governing rights and limitations under Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
{2jtab: CIBC Acknowledgement}
NIH/NIGMS Center for Integrative Biomedical Computing Acknowledgment
CIBC software and the data sets provided on this web site are Open Source software projects that are principally funded through the SCI Institute's NIH/NIGMS CIBC. For us to secure the funding that allows us to continue providing this software, we must have evidence of its utility. Thus we ask users of our software and data to acknowledge us in their publications and inform us of these publications. Please use one of the following acknowledgment and send us references to any publications, presentations, or successful funding applications that make use of the NIH/NIGMS CIBC software or data sets we provide.
"This project was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P41 GM103545 and R24 GM136986.”
