Frequently Asked Questions
- What are the details of the Utah Torso Dataset.
- How do I use relative pathname support in SCIRun 4 network files?
- What is a generic CMake command line for building SCIRun 4?
- What is required to run SCIRun 4?
- Is there a way to automatically quit SCIRun after running a script to completion using the command line?
- I am using ShowFieldGlyphs to visualize tensor data. Colormap Lookup and Conversion to RGB are two Tensor coloring options. With scalar data, understanding the colormap is intuitive. However, with tensors I am unable to understand how the colormap is generated (for both Colormap Lookup and Conversion to RGB options). Is there a mathematical operation performed to convert tensor to scalar?
- How do I take advantage of this built-in Python interpreter?
- I would like to look at the SCIRun field file (.fld) datasets using own visual data analysis software. Is there a description of that format?
- There many files like the SCIRun field file (.fld) in sample datasets (for example, in cyl3). How can I create *.fld file for my own geometry?
- How you place an electrode in order to have it just in contact with the surface of the torso?
- InterfaceWithMatlab fails on Windows because libeng could not be opened. How do I fix this?
- How do I save 3D grid data to a NRRD file that I can use in SCIRun?
- Will SCIRun 4 run on OS X 10.8+?
- Will SCIRun 4 work with the default graphics drivers on Ubuntu 12.10 and newer?
- I need to rotate objects in the SCIRun 4 ViewScene. How do I enable the middle mouse button on Mac OS X?
Datasets
What are the details of the Utah Torso Dataset?
First, the original source of the geometry was MRI, as you have read. However, our interest in the bone data came later, and they were not visible enough in the MRI to extract from there. Instead, we acquired standardized rib and spine geometry and fit them to the torso model.
With that said, there is considerable variation among patients with regard to heart shape and even location. Some hearts are long and relatively narrow while others sit higher and are larger in a transverse slice. The variation you have spotted between the textbook and the Utah torso is, I believe, well within that normal range of variation.
Finally, gating to the R wave does not ensure systole in the images of the heart for several reasons. The first reason is that electrical and mechanical systole are not aligned in time. Each cell of the heart experiences a delay between electrical and mechanical excitation (up to 200 ms in fact). The result is that the R wave occurs during end diastole of the mechanical cycle, i.e., well before mechanical systole. Even more important, using the R wave to gate the image acquisition does not mean that all images have to be captured at the time of the R wave peak; rather the peak of the R wave provides a common reference so that acquisitions from different beats can capture the same phase of the heart cycle. The actual acquisition moment can be offset from the time of the R wave using a simple delay, i.e., each acquisition can occur with the same delay from that peak of the R wave.
I hope that, with this information, you can see that the Utah torso geometry can, indeed, be trusted to reasonably representthe geometry of an adult human torso. Just as with the visible human data, this is only one specific case and there are simply not enough data available to begin to create a true atlas that specifies the ranges of normal anatomy of the entire torso. There are atlases like this of the brain but I don't know of any other organ system that has received enough attention to begin to quantify anatomical variation.
Network Files
We use XML-based .srn (SCIRun Network) files. We strongly suggest you convert your .net files to the new .srn format. If you need any more information, please read the module renaming section under "User Documentation / Modules".
How do I use relative pathname support in SCIRun 4 network files?
The relative path support in the SCIRun network fields assumes the following: The SCIRun network files are located at a fixed relative location from the data. It was designed so you can create a repository in your home directory with data files and networks in the same relative positions that can be easily copied to another directory (another home directory for instance).
You can activate it in your home directory in a file called: .scirunrc
Edit the line:
SCIRUN_NET_RELATIVE_FILENAMES = trueWhen this variable is set all filenames are exported in a relative fashion. Hence, everyone that wants to export to the repository should have this variable set in the .scirunrc filename. Users who only check out files do not need to edit this variable. SCIRun by default reads both files with absolute paths and with relative paths.
To convert files, set the variable in the .scirunrc file. Then load up each network and save the files. This will export all the files in a relative format. Afterwards, check in the files and everything should work.
CMake
What is a generic CMake command line for building SCIRun 4?
The SCIRun directory (which contains the src subdirectory), has a bin directory. In the bin directory, run this command:
cmake ../src
Some other regularly used CMake variables are:
CMAKE_BUILD_TYPE=Debug (or "Release") BUILD_TESTING=ON (or OFF)FAQ
Q: What is required to run SCIRun 4?
A: You can find system requirements for building and running SCIRun under the “Specifications” tab.Q: Is there a way to automatically quit SCIRun after running a script to completion using the command line?
An example using bash in linux would be:
scirun -e test.srn
This would load and execute the SCIRun script test.srn, but I would like to quit/exit it after running.
All you need to do is to change the flag to '-E', so your script would be
scirun -E test.srn
Q: I am using ShowFieldGlyphs to visualize tensor data. Colormap Lookup and Conversion to RGB are two of the Tensor coloring options. With scalar data, understanding the colormap is intuitive. However, with tensors I am unable to understand how the colormap is generated (for both Colormap Lookup and Conversion to RGB options). Is there a mathematical operation performed to convert tensor to scalar?
A: The tensor's principle eigenvector magnitude is used to index to the colormap. The principle eigenvector's x, y, z components are used to generate RGB colors.
Q: How do I take advantage of this built-in Python interpreter?
A: Python scripting support is available in SCIRun 5.
Q: I would like to look at the SCIRun field file (.fld) datasets using my own visual data analysis software. Is there a description of the .fld format?
A: Unfortunately, the SCIRun field file (.fld) format is not well documented. Generally, a SCIRun field file is made up of a small set of tags constituting a header, used to identify the file as a SCIRun field, distinguish between binary and text files, optionally establish endian type etc. The remaining entries are specific to SCIRun's internal data types, identifying mesh types, possible data type contained in the mesh, data location if applicable and other data type-specific information.
Q: There are many files like the SCIRun field file (.fld) in sample datasets (for example, in cyl3). How can I create a .fld file for my own geometry?
A: As you know the SCIRun field file (.fld) is SCIRun's native format. Unfortunately I don't know of any other program that can produce a .fld. My suggestion would be to read in you mesh using the "ReadField" module. There is a drop down menu that allows you to select several different format types. The format will be converted into the "field" format that SCIRun uses. You can then write it out as a .fld with the "WriteField" module.
Q: How do you place an electrode so that it is just in contact with the surface of the torso?
A: This is very much possible, but it is a bit tricky. The idea is that you create a defibrillator source that intersects the torso surface. If you have a paddle geometry (a cube will do to start), you situated it with the EditMeshBoundingBox model so that it intersects the torso (better to be deep than shallow). You then use the ClipFieldByMesh module to just get the intersection of the torso and the paddle. Use GetFieldBoundary to get the boundary of the torso. Find the distance map to the torso boundary in the intersect volume (GetDistanceToField) then clip that field by the distance that you want (2-5 mm) using ClipVolumeByIsovalue. This will be one of your sources in the model. It is not really on the surface but embedded a little beneatch it. This method hasn't been as thoroughly vetted, but it should serve as a starting point.
Q: InterfaceWithMatlab fails on Windows because libeng could not be opened. How do I fix this?
A: This problem has shown up on Windows with recent Matlab releases, and this solution has been tested on Windows 7 64-bit with Matlab R2012b 64-bit. Add the directory containing libeng.dll (should be similar to c:\Program Files\MATLAB\R2012b\bin\win64) to the system PATH.
Q: How do I save 3D grid data to a NRRD file that I can use in SCIRun?
A:The easiest way to create a Teem NRRD is to make two different files. The first is a raw data file (usually .raw), and the second is a NRRD header (.nhdr). There are many details and potential variations in the NRRD file format, but the trick is to be consistent between the header and the data. Provided is a simple example that will work in most cases.
The first step is to write out the data in binary format from Matlab. For simplicity's sake, you will want the data arranged so that the values at each point are listed each point (if there are more than one values per location as with vectors), increasing along the positive x direction, then y direction, then z direction.
For vectors, the values need to be order x, y, z and for tensors the values must be ordered xx, xy, xz, yy, yz, zz of a symmetric tensor matrix. Arrange your data into a 4D matrix in Matlab as you would want it to be in SCIRun (values, x-axis, y-axis,z-axis). Then you write out this data using the fwrite function. This will create a binary file which SCIRun will read in with the help of the header.
A=reshape(data,d1,d2,d3,d4); % reshape the data to 4D matrix of size d1, d2, d3, d4
fid=fopen('data.raw','w','b');
fwrite(fid,A,'float');
fclose(fid);
If you are working with scalar data, d1=1, with vector data, d1=3, and with tensor data, d1=6. For all, d2, d3, and d4 are the number of voxels in the x, y, and z direction respectively.
Note: if you are using scalar data you may reduce the data to a 3D matrix, but you will need to make the corresponding changes to the header file, which will not be discussed here.
The second step of creating a NRRD header can be done using a text editor. You should create a nhdr file (named data.nhdr as an example) as follows:
NRRD0004
type: float
dimension: 4
sizes: d1 d2 d3 d4
encoding: raw
endian: big
datafile: data.raw
space dimension: 3
space origin: (0,0,0)
space directions: none (spacing x,0,0) (0,spacing y,0) (0,0,spacing z)
The origin in this case is the minimum point, and the voxel size is encoded in the space directions (spacing=voxel size).
Notice that the data type in the header matches the type of data in the fwrite function (float) and the format (endian: big) in the header matches the format in the fopen function ('b'). For more information about the NRRD header and file format, see the Teem NRRD file format documentation page.
Once these two files have been created in the same directory, you can use the ReadField module with the NrrdFile (*.nhdr, *.nrrd) option or the ReadNrrd module to read it into SCIRun. If the data volume is oriented strangely or flipped when loaded into SCIRun, usually because of inconsistent coordinate systems, you can rearrange the data in Matlab or change the header file by rearranging the space directions and the size dimension order and flip the axes by making them negative (you may also need to change the origin with flipped axes). The trick here is to make the changes consistent across all the header information.
Teem also offers the command-line unu tool to create and manipulate NRRD files. Finally, questions about the Teem library can be addressed to the teem-users mailing list.
Q: Will SCIRun 4 run on OS X 10.8 (Mountain Lion) and newer versions of OS X?
Testing SCIRun 4 on OS X 10.8 and 10.9 has shown that there is a problem using the ViewScene module on systems with some AMD/ATI graphics cards where the screen will become garbled. The NVIDIA cards we have tested have not shown this problem. AMD FirePro D500 shipped with Mac Pros running OS X Sierra (10.12) and El Capitan (10.11) also appear to be safe to use with XQuartz 2.7.11. This information can also be found under the Specifications and Mac OS X tabs.
Q: Will SCIRun 4 work with the default graphics drivers on Ubuntu 12.10 and newer?
Testing SCIRun 4 on newer versions of Ubuntu (later than 12.04 LTS) has shown that the default graphics drivers are not compatible, i.e. SCIRun cannot run. We test with and recommend NVIDIA cards and drivers, which are known to work.
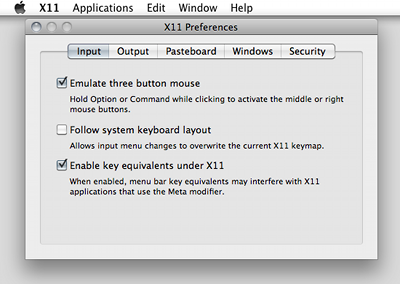
Q: I need to rotate objects in the SCIRun 4 ViewScene. How do I enable the middle mouse button on Mac OS X?
Enable the 'Emulate three button mouse' option in the X11 App Preferences window. X11 App Preferences can be opened from the X11 menu.