2012
M.W. Prastawa, S.P. Awate, G. Gerig.
“Building Spatiotemporal Anatomical Models using Joint 4-D Segmentation, Registration, and Subject-Speci fic Atlas Estimation,” In Proceedings of the 2012 IEEE Mathematical Methods in Biomedical Image Analysis (MMBIA) Conference, pp. 49--56. 2012.
DOI: 10.1109/MMBIA.2012.6164740
PubMed ID: 23568185
PubMed Central ID: PMC3615562
Keywords: namic, adni, autism
R. Ranjan, E.G. Kholmovski, J. Blauer, S. Vijayakumar, N.A. Volland, M.E. Salama, D.L. Parker, R.S. MacLeod, N.F. Marrouche.
“Identification and Acute Targeting of Gaps in Atrial Ablation Lesion Sets Using a Real-Time Magnetic Resonance Imaging System,” In Circulation: Arrhythmia and Electrophysiology, Vol. 5, pp. 1130--1135. 2012.
DOI: 10.1161/CIRCEP.112.973164
PubMed ID: 23071143
PubMed Central ID: PMC3691079
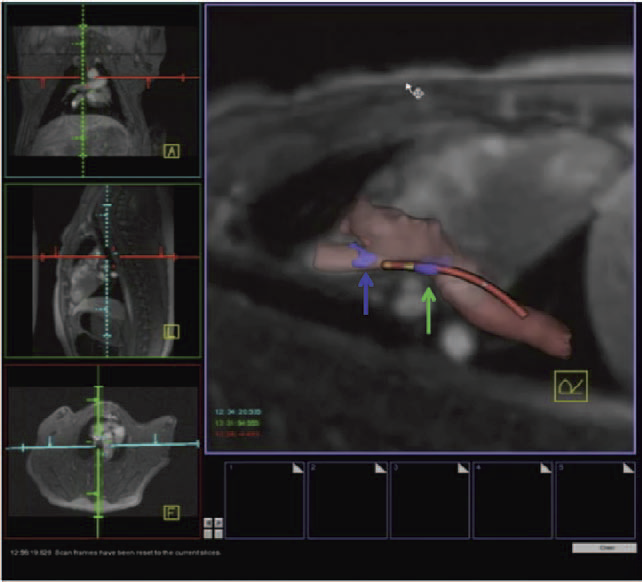
Background - Radiofrequency ablation is routinely used to treat cardiac arrhythmias, but gaps remain in ablation lesion sets because there is no direct visualization of ablation-related changes. In this study, we acutely identify and target gaps using a real-time magnetic resonance imaging (RT-MRI) system, leading to a complete and transmural ablation in the atrium.
Methods and Results - A swine model was used for these studies (n=12). Ablation lesions with a gap were created in the atrium using fluoroscopy and an electroanatomic system in the first group (n=5). The animal was then moved to a 3-tesla MRI system where high-resolution late gadolinium enhancement MRI was used to identify the gap. Using an RT-MRI catheter navigation and visualization system, the gap area was ablated in the MR scanner. In a second group (n=7), ablation lesions with varying gaps in between were created under RT-MRI guidance, and gap lengths determined using late gadolinium enhancement MR images were correlated with gap length measured from gross pathology. Gaps up to 1.0 mm were identified using gross pathology, and gaps up to 1.4 mm were identified using late gadolinium enhancement MRI. Using an RT-MRI system with active catheter navigation gaps can be targeted acutely, leading to lesion sets with no gaps. The correlation coefficient (R2) between the gap length was identified using MRI, and the gross pathology was 0.95.
Conclusions - RT-MRI system can be used to identify and acutely target gaps in atrial ablation lesion sets. Acute targeting of gaps in ablation lesion sets can potentially lead to significant improvement in clinical outcomes.
P. Rosen, V. Popescu.
“Simplification of Node Position Data for Interactive Visualization of Dynamic Datasets,” In IEEE Transactions on Visualization and Computer Graphics (IEEE Visweek 2012 TVCG Track), pp. 1537--1548. 2012.
PubMed ID: 22025753
PubMed Central ID: PMC3411892
P. Rosen.
“Rectilinear Texture Warping for Fast Adaptive Shadow Mapping,” In Proceedings of the ACM SIGGRAPH Symposium on Interactive 3D Graphics and Games (I3D '12), pp. 151--158. 2012.

Conventional shadow mapping relies on uniform sampling for producing hard shadow in an efficient manner. This approach trades image quality in favor of efficiency. A number of approaches improve upon shadow mapping by combining multiple shadow maps or using complex data structures to produce shadow maps with multiple resolutions. By sacrificing some performance, these adaptive methods produce shadows that closely match ground truth.
This paper introduces Rectilinear Texture Warping (RTW) for efficiently generating adaptive shadow maps. RTW images combine the advantages of conventional shadow mapping - a single shadow map, quick construction, and constant time pixel shadow tests, with the primary advantage of adaptive techniques - shadow map resolutions which more closely match those requested by output images. RTW images consist of a conventional texture paired with two 1-D warping maps that form a rectilinear grid defining the variation in sampling rate. The quality of shadows produced with RTW shadow maps of standard resolutions, i.e. 2,048×2,048 texture for 1080p output images, approaches that of raytraced results while low overhead permits rendering at hundreds of frames per second.
Keywords: Rendering, Shadow Algorithms, Adaptive Sampling
L. Zhu, Y. Gao, A. Yezzi, R.S. MacLeod, J. Cates, A. Tannenbaum.
“Automatic Segmentation of the Left Atrium from MRI Images using Salient Feature and Contour Evolution,” In Proceedings of the 34th Annual International Conference of the IEEE EMBS, pp. 3211--214. 2012.
DOI: 10.1109/EMBC.2012.6346648
PubMed ID: 23366609
PubMed Central ID: PMC3652873
2011
N. Akoum, M. Daccarett, C. McGann, N. Segerson, G. Vergara, S. Kuppahally, T. Badger, N. Burgon, T. Haslam, E. Kholmovski, R.S. MacLeod, N.F. Marrouche.
“Atrial fibrosis helps select the appropriate patient and strategy in catheter ablation of atrial fibrillation: a DE-MRI guided approach,” In Journal of Cardiovascular Electrophysiology, Vol. 22, No. 1, pp. 16--22. 2011.
DOI: 10.1111/j.1540-8167.2010.01876.x
PubMed ID: 20807271
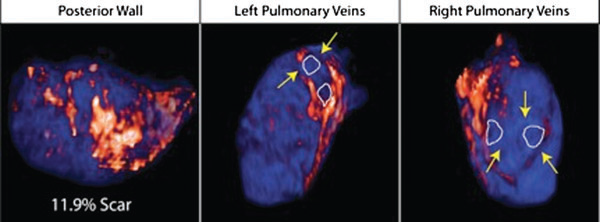
Atrial fibrillation (AF) is the most common sustained arrhythmia encountered in adult cardiology.1,2 Several studies have demonstrated that AF is associated with electrical, contractile, and structural remodeling (SRM) in the left atrium (LA) that contributes to the persistence and sustainability of the arrhythmia.3-7 It has also been shown that the end result of this remodeling process is loss of atrial myocytes and increased collagen content and hence fibrosis of the LA wall.5 Delayed enhancement MRI (DE-MRI) using gadolinium contrast has been demonstrated to localize and quantify the degree of SRM or fibrosis associated with AF in the LA.8
DE-MRI has also been shown to be useful in localizing and quantifying scar formation in the LA following radiofrequency ablation (RFA).9,10 The pulmonary vein (PV) antral region can be visualized to assess circumferential PV scarring that results from RFA lesions/ablation. In addition, the amount of scarring to the LA after catheter ablation can be quantified as a proportion of the total left atrial volume.
Rhythm control of AF using catheter ablation has yielded varying results in different patient populations.11 Identifying the ideal candidate for catheter ablation remains a significant challenge. In addition, a number of different approaches to catheter ablation have been reported and most experts agree that 1 ablation strategy does not fit allAF patients.11-15 Therefore, selecting the proper strategy for a particular patient is also an important determinant of procedure success.
We used DE-MRI to quantify both the degree of SRM/fibrosis pre-ablation and scar formation post ablation. Our aim was to identify predictors of successful ablation in a group of patients stratified according to pre-ablation fibrosis. This would help select the most appropriate ablation strategy for the individual AF ablation candidate.
N. Andrysco, P. Rosen, V. Popescu, B. Benes, K.R. Gurney.
“Experiences in Disseminating Educational Visualizations,” In Lecture Notes in Computer Science (7th International Symposium on Visual Computing), Vol. 2, pp. 239--248. September, 2011.
DOI: 10.1007/978-3-642-24031-7_24
B.M. Burton, J.D. Tate, B. Erem, D.J. Swenson, D.F. Wang, D.H. Brooks, P.M. van Dam, R.S. MacLeod.
“A Toolkit for Forward/Inverse Problems in Electrocardiography within the SCIRun Problem Solving Environment,” In Proceedings of the 2011 IEEE Int. Conf. Engineering and Biology Society (EMBC), pp. 267--270. 2011.
DOI: 10.1109/IEMBS.2011.6090052
PubMed ID: 22254301
PubMed Central ID: PMC3337752
A.N.M. Imroz Choudhury, P. Rosen.
“Abstract Visualization of Runtime Memory Behavior,” In 6th IEEE International Workshop on Visualizing Software for Understanding and Analysis (VISSOFT 2011), pp. 22--29. 2011.

M. Daccarett, T.J. Badger, N. Akoum, N.S. Burgon, C. Mahnkopf, G.R. Vergara, E.G. Kholmovski, C.J. McGann, D.L. Parker, J. Brachmann, R.S. Macleod, N.F. Marrouche.
“Association of left atrial fibrosis detected by delayed-enhancement magnetic resonance imaging and the risk of stroke in patients with atrial fibrillation,” In Journal of the American College of Cardiology, Vol. 57, No. 7, pp. 831--838. 2011.
PubMed ID: 21310320
M. Daccarett, C.J. McGann, N.W. Akoum, R.S. MacLeod, N.F. Marrouche.
“MRI of the left atrium: predicting clinical outcomes in patients with atrial fibrillation,” In Expert Review of Cardiovascular Therapy, Vol. 9, No. 1, pp. 105--111. 2011.
PubMed ID: 21166532
M. Datar, Y. Gur, B. Paniagua, M. Styner, R.T. Whitaker.
“Geometric Correspondence for Ensembles of Nonregular Shapes,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), Lecture Notes in Computer Science (LNCS), Vol. 6892, pp. 368--375. 2011.
DOI: 10.1007/978-3-642-23629-7_45
PubMed ID: 21995050
PubMed Central ID: PMC3346950

Keywords: namic
B. Erem, D.H. Brooks, P.M. van Dam, J.G. Stinstra, R.S. MacLeod.
“Spatiotemporal Estimation of Activation Times of Fractionated ECGs on Complex Heart Surfaces,” In Proceedings of the International Coference of the IEEE Engineering in Medicine and Biology Society (EMBS), pp. 5884--5887. 2011.
DOI: 10.1109/IEMBS.2011.6091455
PubMed ID: 22255678
PubMed Central ID: PMC3345888
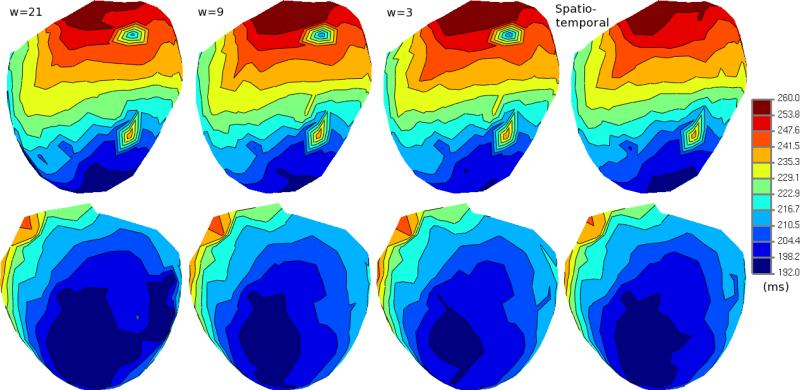
B. Erem, P.M. van Dam, D.H. Brooks.
“Analysis of the Criteria of Activation-Based Inverse Electrocardiography using Convex Optimization,” In Conf Proc IEEE Eng Med Biol Soc, pp. 3913–3916. 2011.
DOI: 10.1109/IEMBS.2011.6090972
PubMed ID: 22255195
PubMed Central ID: PMC3359386
In inverse electrocardiography (ECG), the problem of finding activation times on the heart noninvasively from body surface potentials is typically formulated as a nonlinear least squares optimization problem. Current solutions rely on iterative algorithms which are sensitive to the presence of local minima. As a result, improved initialization approaches for this problem have been of considerable interest. However, in experiments conducted on a subject with Wolff-Parkinson-White syndrome, we have observed that there may be a mismatch between favorable solutions of the optimization problem and solutions with the desired physiological characteristics. In this work, we use a method based on a convex optimization framework to explore the solution space and analyze whether the optimization criteria target their intended objective.
B. Erem, D.H. Brooks.
“Differential Geometric Approximation of the Gradient and Hessian on a Triangulated Manifold,” In Proceeding of the IEEE International Symposium on Biomedical Imaging: from nano to macro, pp. 504--507. 2011.
DOI: 10.1109/ISBI.2011.5872455
PubMed ID: 21712967
PubMed Central ID: PMC3122924
In a number of medical imaging modalities, including measurements or estimates of electrical activity on cortical or cardiac surfaces, it is often useful to estimate spatial derivatives of data on curved anatomical surfaces represented by triangulated meshes. Assuming the triangle vertices are points on a smooth manifold, we derive a method for estimating gradients and Hessians on locally 2D surfaces embedded in 3D directly in the global coordinate system. Accuracy of the method is validated through simulations on both smooth and corrugated surfaces.
T. Fogal, J. Krüger.
“Efficient I/O for Parallel Visualization,” In Proceedings of the Eurographics Symposium on Parallel Graphics and Visualization (2011), Edited by T. Kuhlen and R. Pajarola and K. Zhou, pp. 81--90. 2011.
Z. Fu, W.-K. Jeong, Y. Pan, R.M. Kirby, R.T. Whitaker.
“A fast iterative method for solving the Eikonal equation on triangulated surfaces,” In SIAM Journal of Scientific Computing, Vol. 33, No. 5, pp. 2468--2488. 2011.
DOI: 10.1137/100788951
PubMed Central ID: PMC3360588
This paper presents an efficient, fine-grained parallel algorithm for solving the Eikonal equation on triangular meshes. The Eikonal equation, and the broader class of Hamilton–Jacobi equations to which it belongs, have a wide range of applications from geometric optics and seismology to biological modeling and analysis of geometry and images. The ability to solve such equations accurately and efficiently provides new capabilities for exploring and visualizing parameter spaces and for solving inverse problems that rely on such equations in the forward model. Efficient solvers on state-of-the-art, parallel architectures require new algorithms that are not, in many cases, optimal, but are better suited to synchronous updates of the solution. In previous work [W. K. Jeong and R. T. Whitaker, SIAM J. Sci. Comput., 30 (2008), pp. 2512–2534], the authors proposed the fast iterative method (FIM) to efficiently solve the Eikonal equation on regular grids. In this paper we extend the fast iterative method to solve Eikonal equations efficiently on triangulated domains on the CPU and on parallel architectures, including graphics processors. We propose a new local update scheme that provides solutions of first-order accuracy for both architectures. We also propose a novel triangle-based update scheme and its corresponding data structure for efficient irregular data mapping to parallel single-instruction multiple-data (SIMD) processors. We provide detailed descriptions of the implementations on a single CPU, a multicore CPU with shared memory, and SIMD architectures with comparative results against state-of-the-art Eikonal solvers.
B.M. Isaacson, J.G. Stinstra, R.D. Bloebaum, COL P.F. Pasquina, R.S. MacLeod.
“Establishing Multiscale Models for Simulating Whole Limb Estimates of Electric Fields for Osseointegrated Implants,” In IEEE Transactions on Biomedical Engineering, Vol. 58, No. 10, pp. 2991--2994. 2011.
DOI: 10.1109/TBME.2011.2160722
PubMed ID: 21712151
PubMed Central ID: PMC3179554
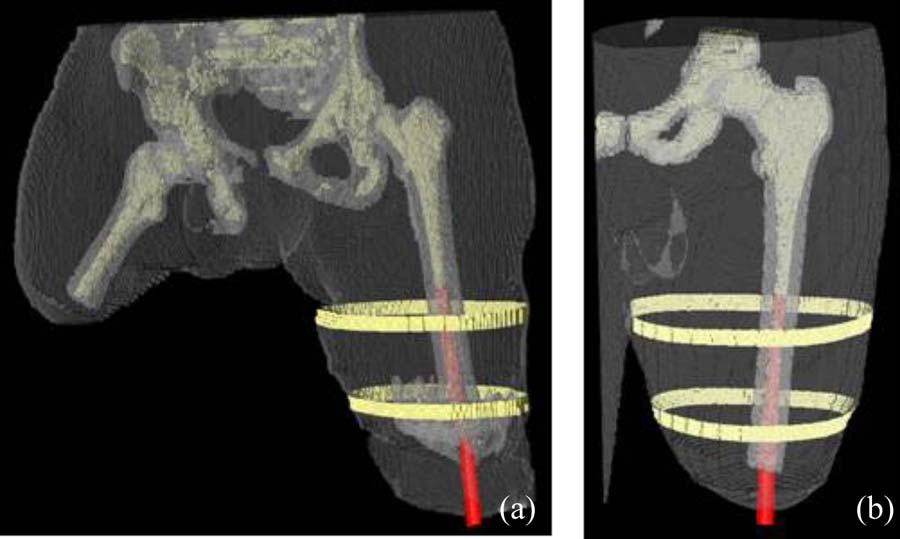
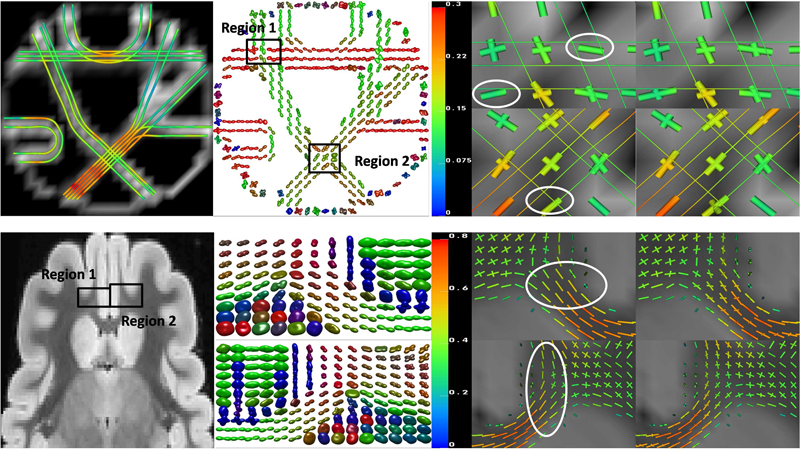
F. Jiao, Y. Gur, C.R. Johnson, S. Joshi.
“Detection of crossing white matter fibers with high-order tensors and rank-k decompositions,” In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI 2011), Lecture Notes in Computer Science (LNCS), Vol. 6801, pp. 538--549. 2011.
DOI: 10.1007/978-3-642-22092-0_44
PubMed Central ID: PMC3327305
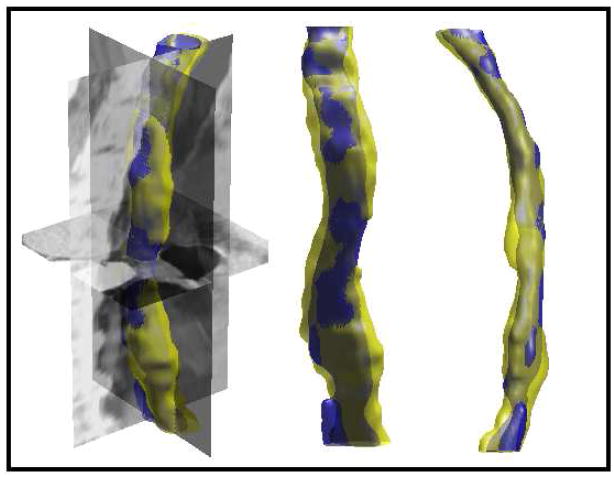
S. Kurugol, E. Bas, D. Erdogmus, J.G. Dy, G.C. Sharp, D.H. Brooks.
“Centerline extraction with principal curve tracing to improve 3D level set esophagus segmentation in CT images,” In Proceedings of IEEE International Conference of the Engineering in Medicine and Biology Society (EMBS), pp. 3403--3406. 2011.
DOI: 10.1109/IEMBS.2011.6090921
PubMed ID: 2225507
PubMed Central ID: PMC3349355
Page 10 of 24