Electron Microscopy Segmentation
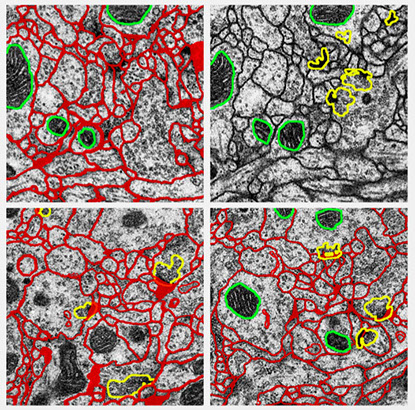
Segmentation of Electron Microscopy images is one of the main aspects of research in our group. We developed and proposed several different methods to address this problem. Accurately segmentation of individual neuron cells in electron microscopy (EM) images is very important to quantitative connectomics research. Electron microscopy images of neural tissue are typically hard to segment due to their properties. The staining used in electron microscopy is non-selective, i.e. it highlights all cellular and intracellular membranes present in the tissue sample. Therefore, a successful segmentation of cells first requires the differentiation of cellular membranes from intracellular membranes, which look similar at the local scale but are differentiable using non-local contextual cues. The first step for such segmentation is to automatically detecting cell membranes using supervised machine learning, which results in probability maps of cell boundaries. Then region segmentation is generated that assigns each neuron cell a unique integer label. We propose CHM as a state-of-the-art probability map generator and HMT as a state-of-the-art region segmentation method.
Cascaded Hierarchical Model (CHM) is a multi-resolution contextual framework and learns contextual information in a hierarchical framework for image segmentation. CHM, combined with LDNN achieves state-of-the-art on segmentation of different structures in EM images.
HMT is a fully automatic approach for intra-section segmentation and inter-section reconstruction of neurons in Electron Microscopy images by building a hierarchical merge tree structure to represent multiple region hypotheses
