Particle-based Shape Modeling
Motivation: Statistical modeling of shape has already proven useful in many areas of medicine where recent advances in medical imaging have made shape modeling feasible for the study of form and function of different anatomical structures with higher throughput and less human intervention. Shape is the characteristic that remains after removing all global geometrical information from an object. To study shape, we would like to study the differences among these characteristics in populations of objects belonging to the same class. It turned out that analyzing shapes can answer various questions imposed by different applications. For example, shape analysis can quantify the developmental changes over time as a response for a gene mutation. Further, the study of shape can boost classification, for example in biology, to discriminate different species based on the shape of their bones. In neuroanatomy, shape differences can help diagnosing schizophrenic people through the study of the shape of brain structures. In biomechanics, we can quantify the shape changes of certain joints as function of age.
A core machinery to study shape is statistics. Statistics is an important branch of mathematics, and is widely used to study/characterize data (in our case, shapes). For example, regression analysis is used to study the change of dependent processes w.r.t an explanatory variable. Statistics can also quantify group differences between healthy and pathological groups. Such quantification could help doctors to diagnose and probably stage the severity of a specific disease in order to tailor his/her treatment plan towards a specific patient.
To perform statistics on shapes, we need to come up with a compact, yet accurate representation of such a shape. One way of doing this is to represent each shape as an ordered set of points on which we can compute statistics in the space defined these sets. However, in order to have a meaningful shape statistics that reflect variability among the given population, these sets of points need to be placed in corresponding locations for each shape. For instance, for a hand’s shape, we can choose the tips of the fingers to represent such a shape. This might be a compact representation that would exist for each normal hand. But it is not enough to model all geometric details of such a hand. The question now becomes how could we choose the same points across different instances in the same population so that this representation is compact while capturing enough geometric details of the underlying shapes.
- ShapeWorks: particle-based shape correspondence and visualization software
- ShapeWorks for quantifying the spectrum of hip joint pathology
- ShapeWorks for characterizing scapular morphology in Hill-Sachs patients
- Dense particle-based correspondence models and surface reconstruction
- Learning deep features for correspondence optimization
- Estimating shapes directly from images
- ShapeWorks for modeling cranial nerves
ShapeWorks: particle-based shape correspondence and visualization software
Joint work with: Ross Whitaker and Joshua Cates
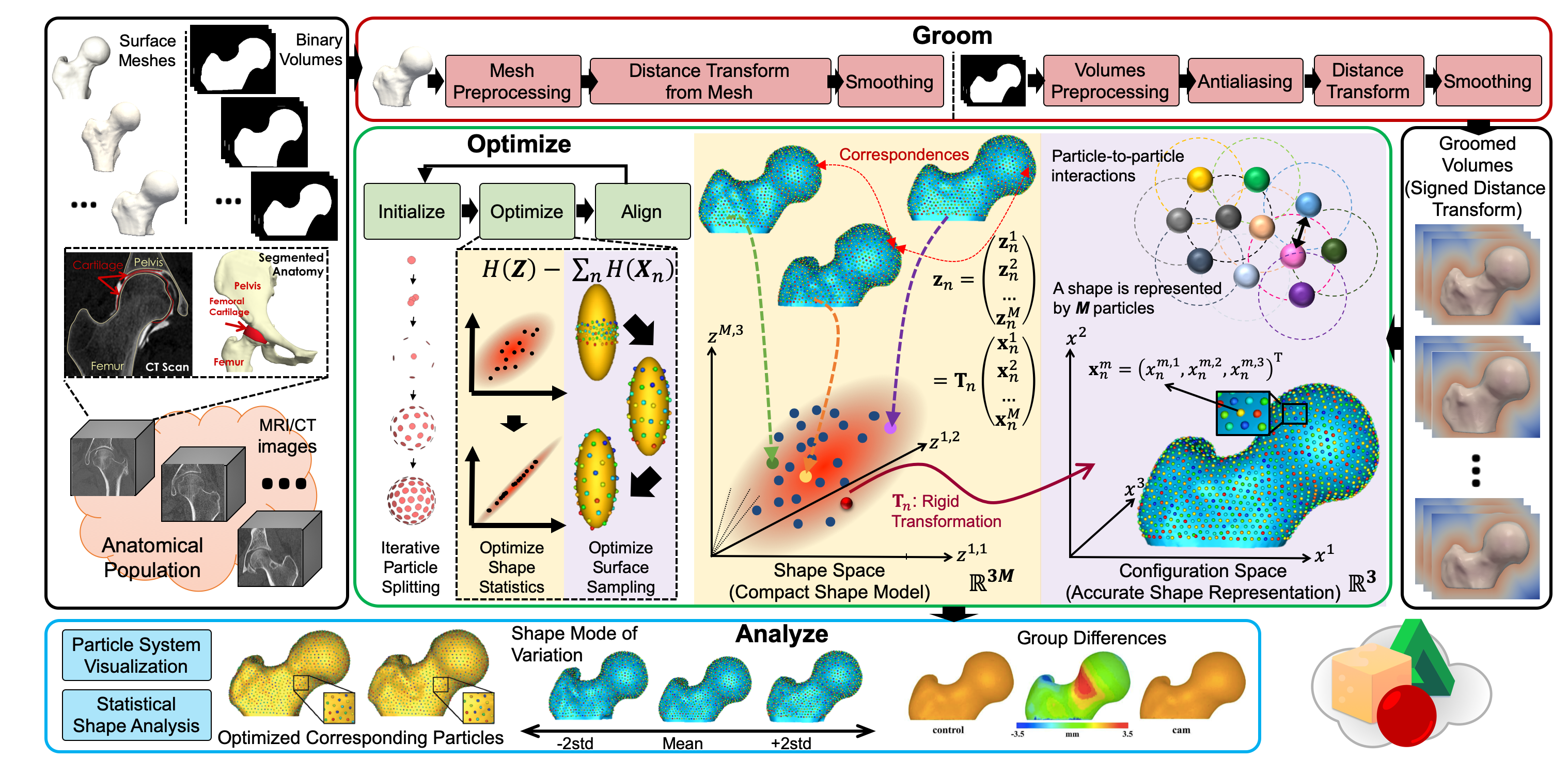
One of the challenges with the applications of statistical shape modeling is that the results from the model depend heavily on how one places the landmarks. My work builds on the prior work of particle-based modeling (PBM) framework, in which the correspondences across a population of shapes are optimized to account for the structure of the population as a whole. The placement of correspondence points is treated as an energy minimization problem, which iteratively moves correspondence points on the surfaces to find a compromise between regular placements of landmarks and landmarks that provide the simplest description of the resulting ensemble statistics. My efforts in this area focus on the extended development and evaluation of particle-based shape modeling (as an explicit shape representation) that is currently implemented in the ShapeWorks software package.
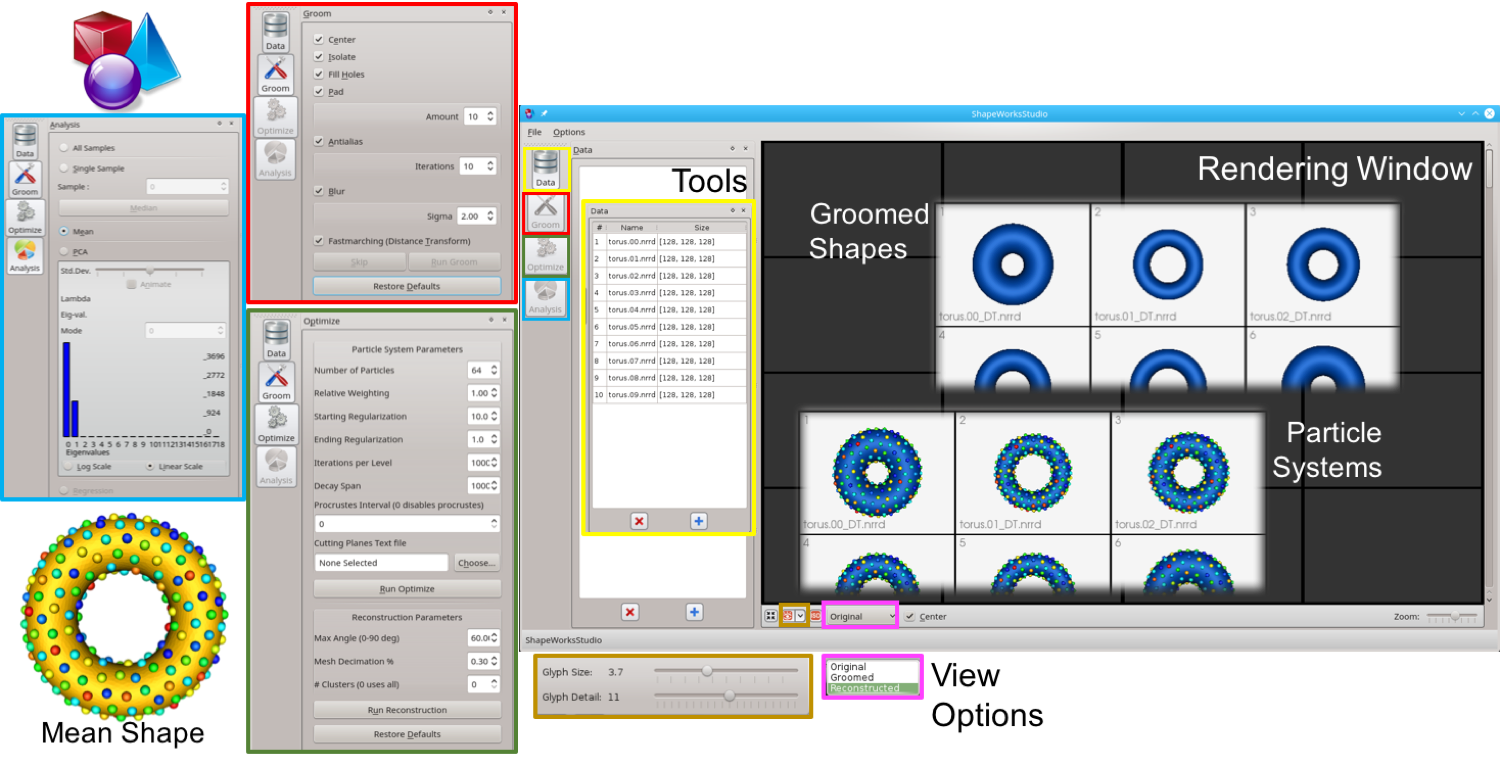
ShapeWorks is an open-source software that enable the automated placement of dense corresponding landmarks on a group of shapes given as binary volumes. ShapeWorks includes preprocessing tools for binary segmentations in order to move particles around in a numerically stable way. ShapeWorks includes the core components of computing landmark-based shape models. It also includes a user interface to analyze and visualize the optimized shape models.
ShapeWorks is the only publicly available tool that learns – by virtue of its automated correspondence mechanism – a population-specific metric in a way that does not penalize natural variability and therefore can capture the underlying parameters in an anatomical shape space. As a demonstration, consider an ensemble of 3D boxes with a bump at a varying location. When comparing the shape of different boxes, one would want to downplay the impact of the bump location on the comparing metric to respect the natural population variability that is not captured by simple affine transformations. Here we illustrate how ShapeWorks is able to discover the underlying mode of variation in the box-bump ensemble in comparison to existing publicly available shape modeling software packages.
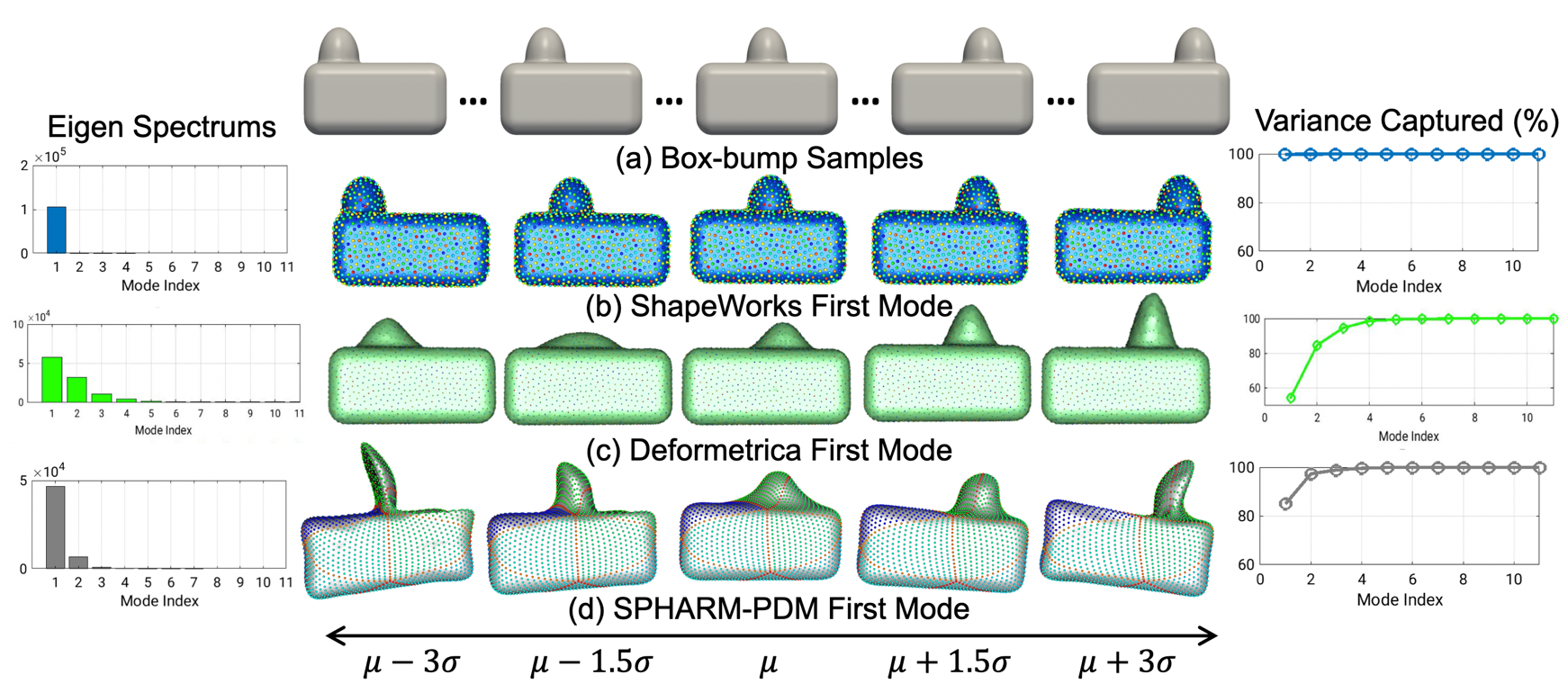
(a) Box-bump samples. The mean \(\pm\) 3 stds of the 1st mode of (b) ShapeWorks (groupwise, parameterization-free, and topology-independent) versus (c) Deformetrica [1], and (d) SPHARM-PDM [2] (pairwise, parameterized, and spherical topology).
[1] S. Durrleman, M. Prastawa, N. Charon, J. R. Korenberg, S. Joshi,
G. Gerig, and A. Trouve. Morphometry of anatomical shape complexes with
dense deformations and sparse parameters. NeuroImage, 101(0):35 – 49,
2014.
[2] M. Styner, I. Oguz, S. Xu, C. Brechbühler, D. Pantazis, J. J.
Levitt, M. E. Shenton, and G. Gerig. Framework for the statistical shape
analysis of brain structures using SPHARM-PDM. The Insight Journal,
(1071):242, 2006.
ShapeWorks also includes a full graphical user interface called ShapeWorksStudio, which can be used to interactively run all of the steps in the shape modeling pipeline. All ShapeWorks source code, binaries, and user documentation are freely available from the ShapeWorks website under the MIT license that is General Public License (GPL) compatible.
Recently, the increased availability of high-resolution in vivo images of anatomy has led to the development and distribution of open-source computational tools to model anatomical shapes and their variability within populations with unprecedented detail and statistical power. Nonetheless, there is little work on the evaluation and validation of such tools as related to clinical applications that rely on morphometric quantifications for treatment planning. To address this lack of validation, we systematically assess the outcome of widely used off-the-shelf SSM tools, namely ShapeWorks, SPHARM-PDM, and Deformetrica, in the context of designing closure devices for left atrium appendage (LAA) in atrial fibrillation (AF) patients to prevent stroke, where an incomplete LAA closure may be worse than no closure. This study is motivated by the potential role of SSM in the geometric design of closure devices, which could be informed by population-level statistics, and patient-specific device selection, which is driven by anatomical measurements that could be automated by relating patient-level anatomy to population-level morphometrics. Hence, understanding the consequences of different SSM tools for the final analysis is critical for the careful choice of the tool to be deployed in real clinical scenarios. Results demonstrate that estimated measurements from ShapeWorks model are more consistent compared to models from Deformetrica and SPHARM-PDM. Furthermore, ShapeWorks and Deformetrica shape models capture clinically relevant population-level variability compared to SPHARM-PDM models.
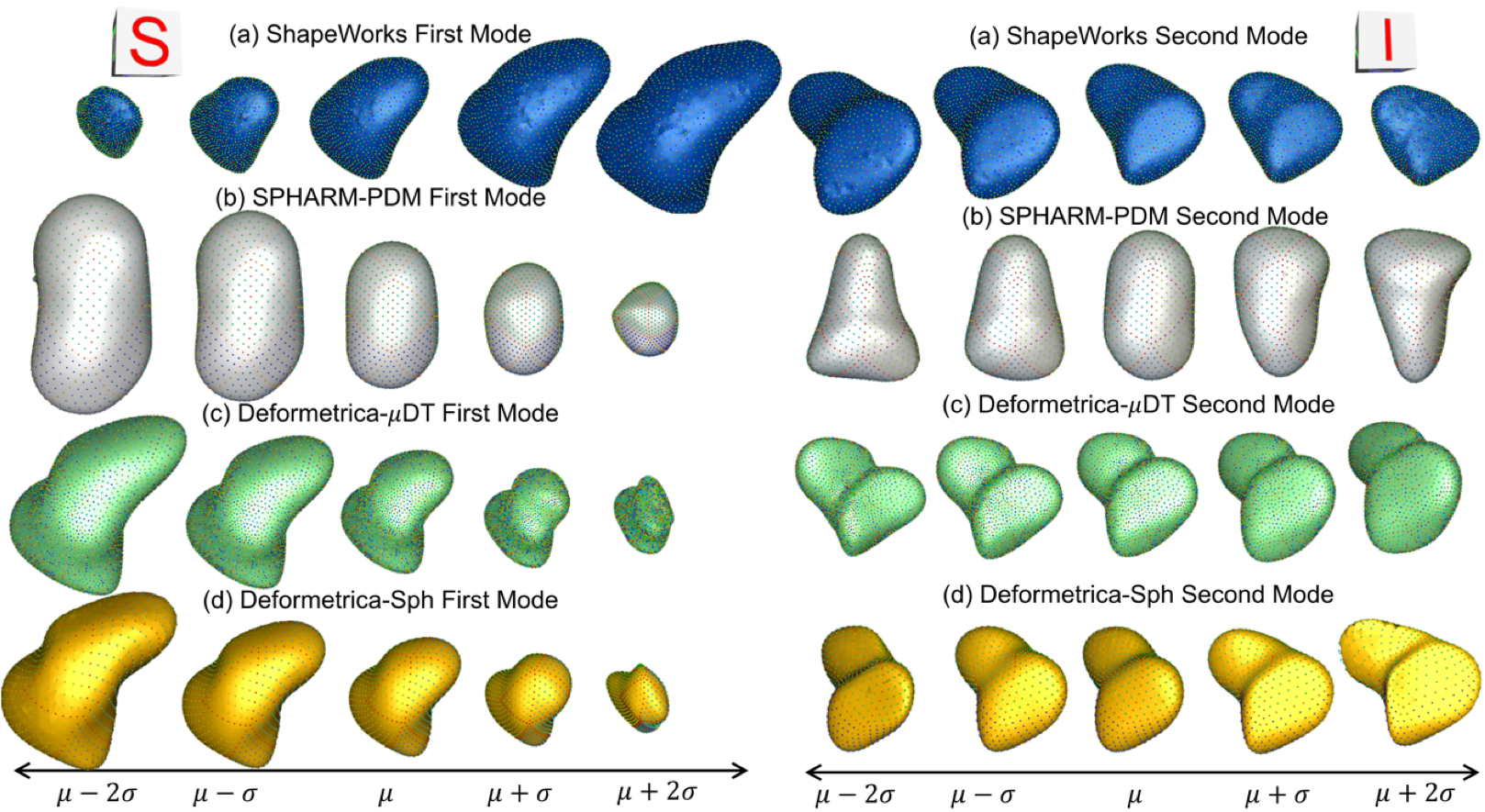
The mean \(\pm\) 2 stds of the 1st and 2nd mode of LAA shape models from (a) ShapeWorks, (b) SPHARM-PDM, (c) Deformetrica-\(\mu\)DT (mean LAA distance transform as the atlas), and (d) Deformetrica-Sph (sphere shape as the atlas). The clinically relevant modes of variation are first mode - elongation of the appendage, second mode - size of the LAA ostia. Compared to other SSM tools, ShapeWorks captured clinically relevant modes of variation.
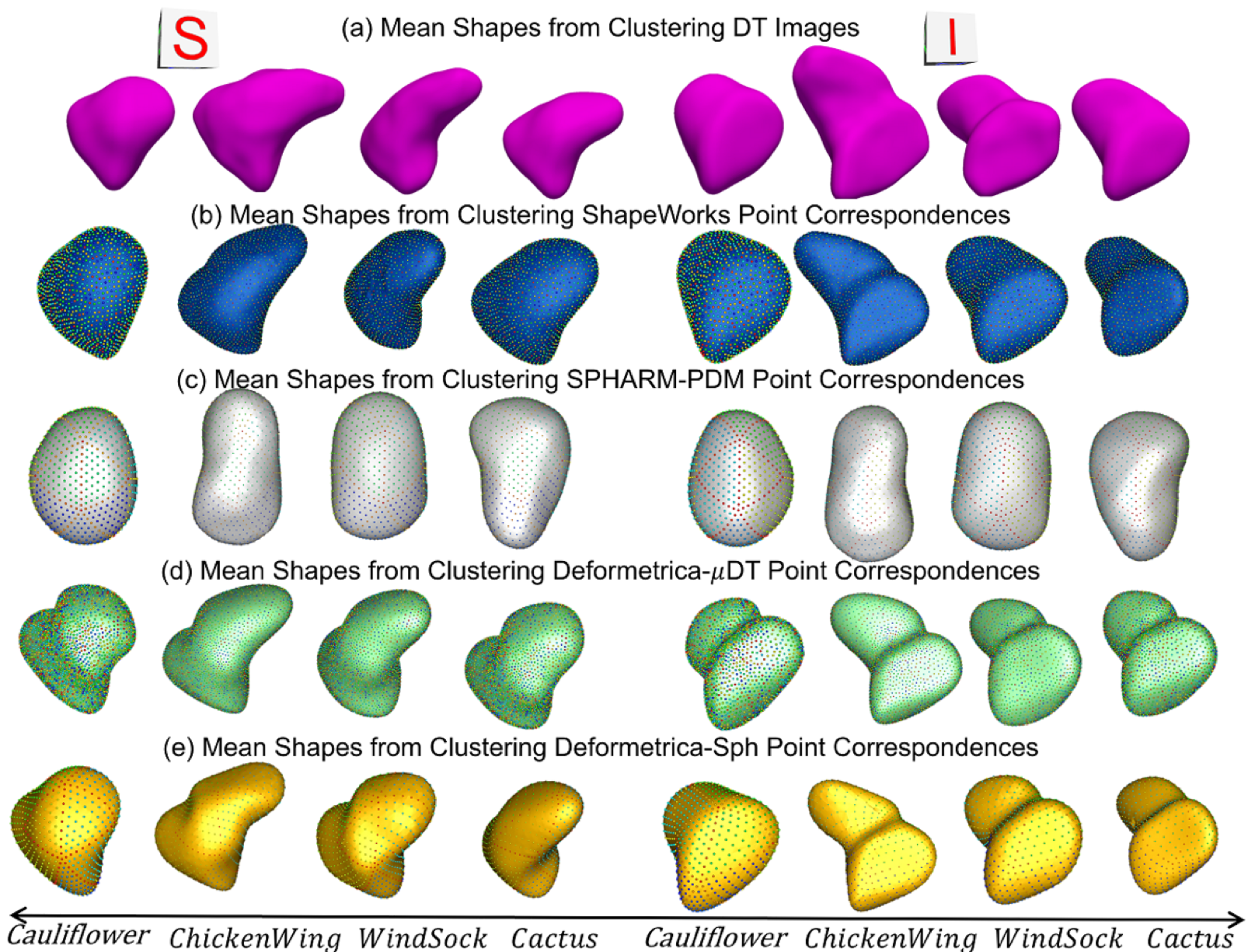
The mean shapes from k-means clustering from (a) Distance Transform (DT) images, (b) ShapeWorks, (c) SPHARM-PDM, (d) Deformetrica-\(\mu\)DT (mean LAA distance transform as the atlas), and (e) Deformetrica-Sph (sphere shape as the atlas). Cluster centers from ShapeWorks and Deformetrica shape models are closely aligned with the centers from distance transforms.
Related publications:
Joshua Cates, Shireen Y. Elhabian, Ross T. Whitaker. ShapeWorks: Particle-based shape correspondence and visualization software. In Statistical Shape and Deformation Analysis: Methods, Implementation and Applications. 1st Edition, Academic Press, 2017.
Shireen Y. Elhabian, Praful Agrawal, Joshua Cates, Manasi Datar, Ross Whitaker. ShapeWorksStudio: Particle-based Shape Correspondence and Visualization Software. Technical Report. Scientific Computing and Imaging Institute, University of Utah, July 2017
Anupama Goparaju, Ibolya Csecs, Alan Morris, Evgueni Kholmovski, Nassir Marrouche, Ross T. Whitaker, and Shireen Y. Elhabian. On the Evaluation and Validation of Off-the-shelf Statistical Shape Modeling Tools: A Clinical Application. ShapeMI-MICCAI 2018: Workshop on Shape in Medical Imaging, 2018.
ShapeWorks for quantifying the spectrum of hip joint pathology
Joint work with: Ross Whitaker, Andrew Anderson, Jeff Weiss, and Christopher Peters
One particular application for ShapeWorks is quantifying the spectrum of hip joint pathology. Femoroacetabular impingement (FAI) is a recently described disease of the hip joint and has received an extraordinary amount of clinical and research attention in recent years. FAI is marked by reduced clearance between the femoral head and acetabulum due to morphologic abnormalities of the femur, acetabulum , or both. This causes some portion of the soft tissue around the hip socket to get pinched or compressed.
Cam-type impingement (more common in men), in particular, occurs when the round head of the femur is not as round as it should be to move properly inside the hip socket. The treatment for this type of pathology is surgical debridement. If physicians could be provided with knowledge of the size and shape of a cam lesion, a more exact diagnosis, as well as detailed preoperative planning, may be facilitated.
ShapeWorks has been able to quantify differences in anatomy and mean cortical thickness for cam and control groups. With statistical models built by ShapeWorks, the cam lesion was detected as the region of the head–neck junction with greater than 1.5mm of difference in the anatomy. Further, ShapeWorks has been able to determine a linear discrimination of the variation between the mean cam and mean control shapes. With a shape score that depends on group-specific mean shapes, ShapeWorks can place a subject-specific anatomy on a disease spectrum that is statistically derived from the shape population. This would provide a physician an objective and data-driven scoring system to stage the disease severity of a patient in order to tailor his/her treatment plan towards this patient.
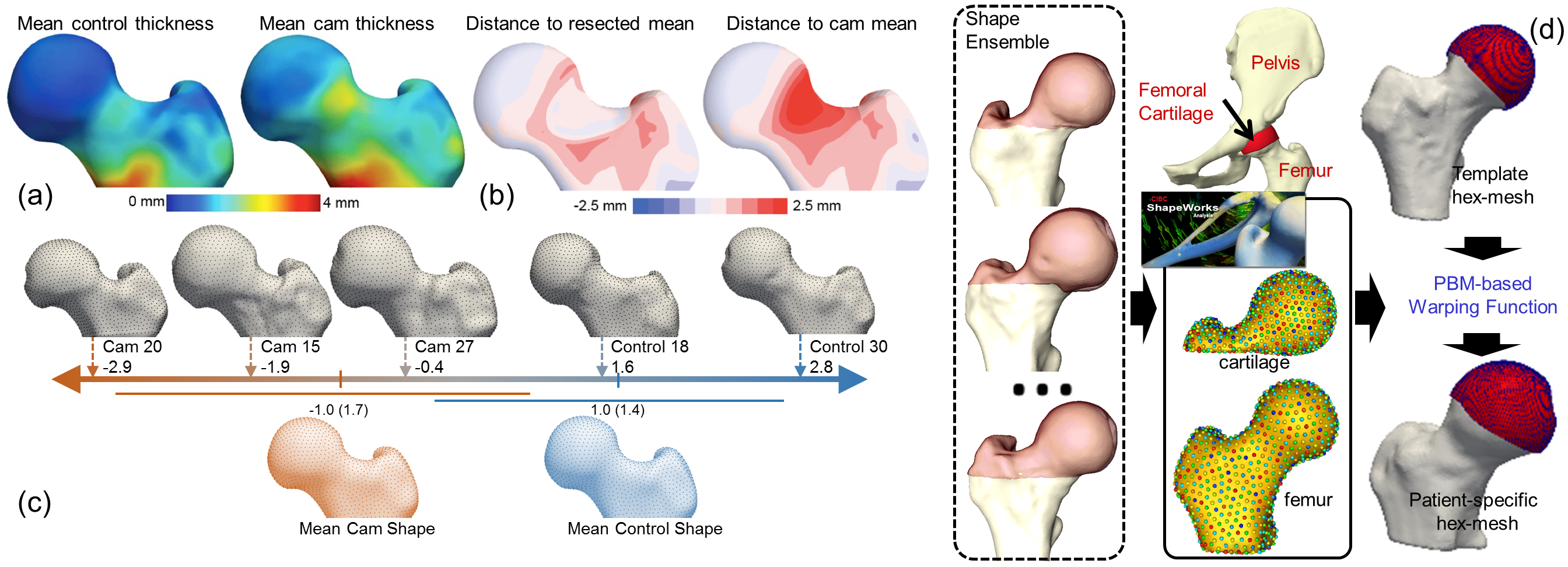
ShapeWorks in orthopedics: (a) Mean cortical thickness of the control and cam-FAI groups in which the greatest thickness difference corresponded to the cam lesion. (b) A simulated resection of the full cortex thickness in the lesion region. The mean shapes for the resected cam and native cam were compared to the mean control shape. (c) Linear discrimination between the cam-FAI and control shapes in shape space. (d) ShapeWorks correspondence model is used to warp a hexahedral mesh from template subject space to every subject.
Related publications:
Penny R. Atkins, Shireen Y. Elhabian, Praful Agrawal, Michael D. Harris, Ross T. Whitaker, Jeffrey A. Weiss, Christopher L. Peters, Andrew E. Anderson. Quantitative Comparison of Cortical Bone Thickness Using Correspondence-based Shape Modeling in Patients With Cam Femoroacetabular Impingement. Journal of Orthopaedic Research (2016).
Anderson, Andrew, Penny R. Atkins, Praful Agrawal, Shireen Y. Elhabian, Ross T. Whitaker, Jeffrey A. Weiss, Christopher L. Peters, Stephen Kenji Aoki. Which Radiographic Measurements Best Identify Anatomical Variation in Femoral Head Anatomy? Analysis Using 3D Computed Tomography and Statistical Shape Modeling. Journal of Hip Preservation Surgery, 3, no. suppl 1 (2016).
Penny Atkins, Stephen Aoki, Shireen Y. Elhabian, Praful Agrawal, Ross Whitaker, Jeffrey Weiss, Christopher Peters, Andrew Anderson. Evaluation of the Sclerotic Subchondral Bone Boundary as a Surgical Resection Guide in the Treatment of Cam-type Femoroacetabular Impingement. Abstract for poster presentation for Annual Meeting of Orthopaedic Research Society, 2017.
Penny Atkins, Praful Agrawal, Shireen Y. Elhabian, Ross Whitaker, Jeffrey Weiss, Christopher Peters, Stephen Aoki, Andrew Anderson. Which Radiographic Measurements Best Identify Anatomical Variation In Femoral Head Anatomy? Analysis Using 3D Computed Tomography and Statistical Shape Modeling. Abstract for podium presentation at Annual Scientific Meeting of International Society of Hip Arthroscopy, 2016, San Francisco, CA.
Penny Atkins, Prateep Mukherjee, Shireen Y. Elhabian, Sumedha Singla, Michael Harris, Jeffrey Weiss, Ross Whitaker, Andrew Anderson. Proximal Femoral Cortical Bone Thickness in Patients with Femoroacetabular Impingement and Normal Hips Analyzed Using Statistical Shape Modeling. Abstract for podium presentation, Summer Biomechanics, Bioengineering and Biotransport Conference, 2015, Snowbird, UT.
Penny Atkins, Prateep Mukherjee, Shireen Y. Elhabian, Sumedha Singla, Michael Harris, Ross Whitaker, Jeffrey Weiss, Andrew Anderson. Warping of Template Meshes for Efficient Subject Specific FE Mesh Generation. Abstract for podium presentation, International Symposium on Computer Methods in Biomechanics and Biomedical Engineering, 2015. Montreal, Canada.
Penny Atkins, Prateep Mukherjee, Sumedha Singla, Shireen Y. Elhabian, Michael Harris, Jeffrey Weiss, Ross Whitaker, Andrew Anderson. Statistical Shape Modeling to Quantify and Comparison of Proximal Femoral Cortical Bone Thickness between Patients with Femoroacetabular Impingement and Normal Hips Analyzed by Statistical Shape Modeling. Abstract for podium presentation, Orthopaedic Research Society, 2015, Las Vegas, NV.
Penny Atkins, Prateep Mukherjee, Sumedha Singla, Shireen Y. Elhabian, Michael Harris, Jeffrey Weiss, Ross Whitaker, Andrew Anderson. Statistical Shape Modeling of Cortical Bone Thickness in Patients with Femoroacetabular Impingement. Abstract for podium presentation. Biomedical Engineering Society, 2014, San Antonio, TX.
ShapeWorks for characterizing scapular morphology in Hill-Sachs patients
Joint work with: Heath B. Henninger and Robert Z. Tashjian
Hill-Sachs lesions are defined as a compression fracture resulting from impaction of the soft cancellous bone of the humeral head against the anterior rim of the glenoid during an anterior dislocation event. These defects are an important risk factor for the recurrence of instability and are a marker for significant instability of the shoulder. In this work, we aim at quantifying scapular morphology between subjects with Hill-Sachs lesions and controls. These efforts will ultimately help in developing a thorough understanding of the association between Hill-Sachs lesions and premorbid morphologic variations and the risk of anterior glenohumeral instability. In this regard, ShapeWorks has been shown to be a powerful tool to compare complex morphology without idealizing underlying geometry, and identify unknown morphologic differences of the scapula. In particular, previously unknown shape variations of the coracoid and acromion, as well as glenoid morphology differences, were identified in scapulae with anterior shoulder instability.
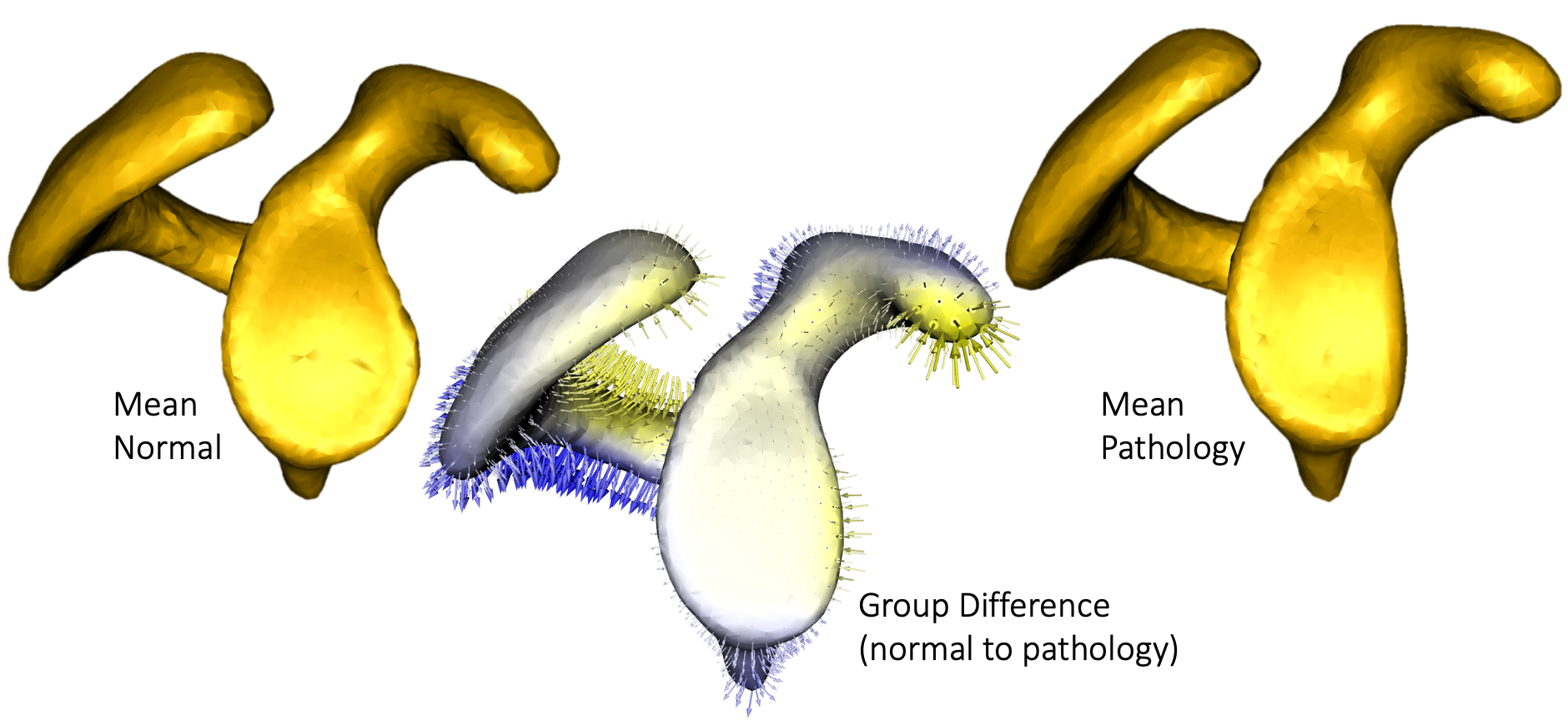
Mean scapula shapes from control and pathologic populations along with group differences.
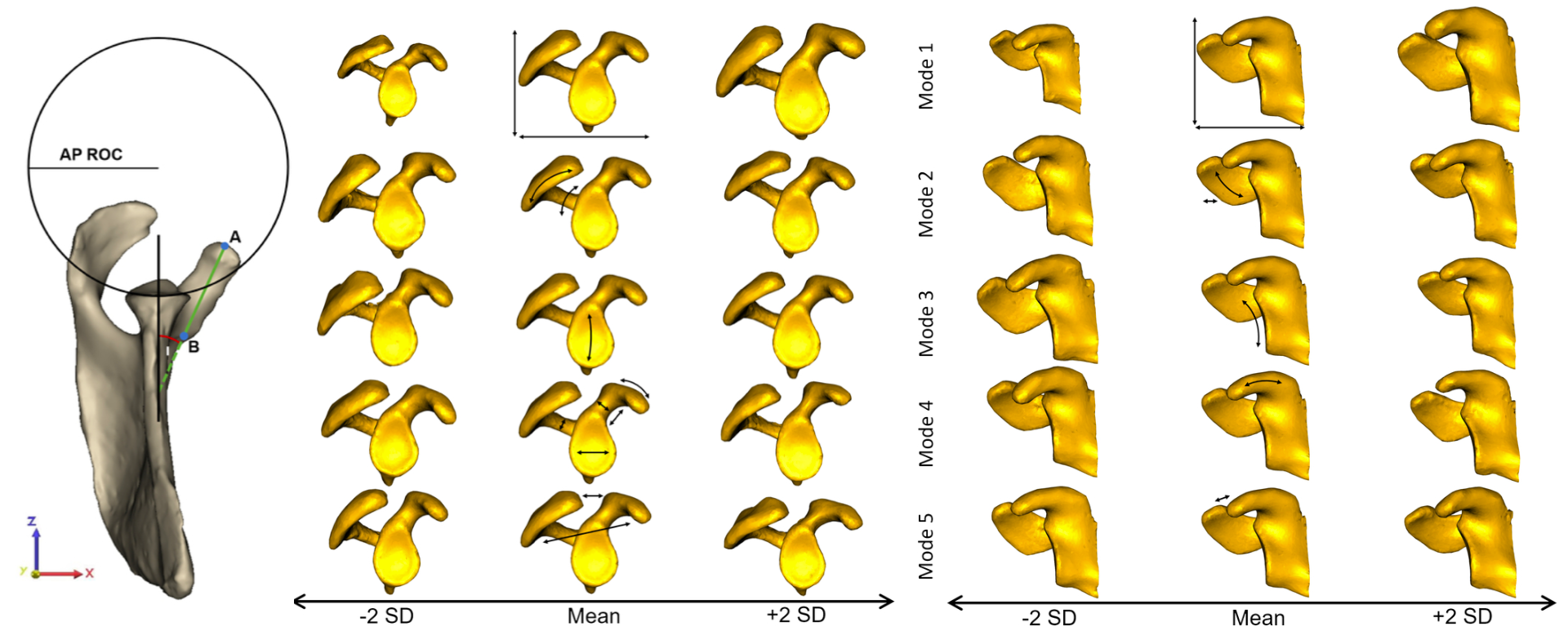
The first 5 modes of variation, (left) illustrated in the plane of the glenoid and (right) in the coronal plane, represented a significant difference in shape, capturing 89.7% of variation. Shapes are presented at ±2 SD with respect to the mean. Arrows on the mean shape highlight the areas of greatest variation captured by each mode. Mode 1 (33.0% of variation) represented scaling differences. Mode 2 (32.0% of variation) demonstrated large differences around the acromion. In Mode 3 (11.8% of variation) variation in glenoid inclination and concavity of the glenoid surface was the most substantial. Mode 4 (9.0% of variation) captured primarily differences in orientation of the coracoid pillar, coracoid process size and bony prominence. Variation in deviation of the coracoid process and the resulting coracoacromial relationship were captured in Mode 5 (3.1% of variation).
Related publications:
Matthijs Jacxsens, Shireen Y. Elhabian, Sarah Brady, Peter Chalmers, Andreas Mueller, Robert Tashjian, Heath Henninger. Thinking outside the glenohumeral box: Hierarchical shape variation of the periarticular anatomy of the scapula using statistical shape modeling. Journal of Orthopaedic Research, in press, 2020.
Matthijs Jacxsens, Shireen Y. Elhabian, Sarah Brady,
Peter Chalmers, Robert Tashjian, Heath Henninger. Coracoacromial
Morphology: A Contributor to Recurrent Traumatic Anterior Glenohumeral
Instability?. Journal of Shoulder and Elbow Surgery, 28(7),
pp. 1316-1325, 2019.
Matthijs Jacxsens, Shireen Y. Elhabian, Robert Z. Tashjian1, Heath B. Henninger. Scapular Morphology In Patients With Hill-Sachs Lesions Using Statistical Shape Modeling. Abstract for podium presentation for the 27th Congress of the European Society for Surgery of the Shoulder and the Elbow (SECEC-ESSSE) conference, 2017.
Dense particle-based correspondence models and surface reconstruction
Joint work with: Ross Whitaker and Joshua Cates
Shapeworks yields relatively sparse correspondence models that may be inadequate to reconstruct thin structures and high curvature regions of the underlying anatomical surfaces. However, for many applications, we require a denser correspondence model, for example, to construct better surface meshes, make more detailed measurements, or conduct biomechanical or other simulations on mesh surfaces. One option for denser modeling is to increase the number of particles per shape sample. However, this approach necessarily increases the computational overhead, especially when modeling large clinical cohorts.
Here we adopt a template-deformation approach to establish an inter-sample dense surface correspondence, given a sparse set of optimized particles. To avoid introducing bias due to the template choice, we propose an unbiased framework for template mesh construction. The dense template mesh is then constructed by triangulating the isosurface of the mean distance transform. This unbiased strategy will preserve the topology of the desired anatomy by taking into account the shape population of interest. In order to recover a sample-specific surface mesh, a warping function is constructed using the sample-level particle system and the mean/template particle system as control points. This warping function is then used to deform the template dense mesh to the sample space.
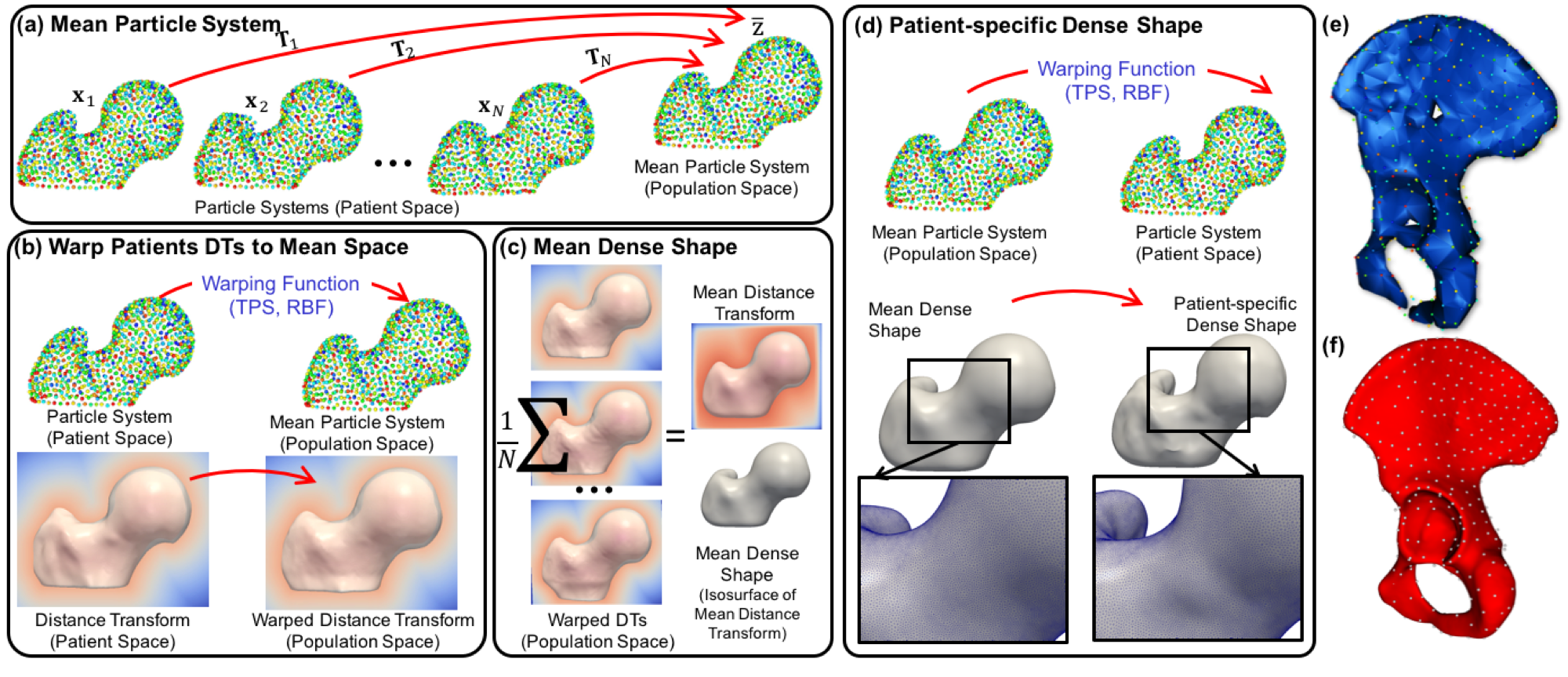
(a–d) Proposed particle-based surface reconstruction. (e,f) A pelvis surface reconstruction with an increased number of particles in which poor reconstructions are observed with smaller number of particles, especially along the pubic arch and iliac crest. Hence, an extension is needed to recover anatomically plausible and accurate 3D shapes at both the population and sample levels from a sparse set of particles.
Related publications:
Joshua Cates, Shireen Y. Elhabian, Ross T. Whitaker. ShapeWorks: Particle-based shape correspondence and visualization software. In Statistical Shape and Deformation Analysis: Methods, Implementation and Applications. 1st Edition, Academic Press, 2017.
Shireen Y. Elhabian, Praful Agrawal, Joshua Cates, Manasi Datar, Ross Whitaker. ShapeWorksStudio: Particle-based Shape Correspondence and Visualization Software. Technical Report. Scientific Computing and Imaging Institute, University of Utah, July 2017
Penny Atkins, Prateep Mukherjee, Shireen Y. Elhabian, Sumedha Singla, Michael Harris, Ross Whitaker, Jeffrey Weiss, Andrew Anderson. Warping of Template Meshes for Efficient Subject Specific FE Mesh Generation. Abstract for podium presentation, International Symposium on Computer Methods in Biomechanics and Biomedical Engineering, 2015. Montreal, Canada.
Learning deep features for correspondence optimization
Joint work with: Ross Whitaker
Currently, particle-based modeling (PBM) relies on a Gaussian assumption in the shape space, which is the vector space formed by the spatial coordinates of surface correspondences (modulo a similarity transform). However, the distribution of anatomical structures can be far more complex than the Gaussian assumes. To address this, previous work (in the context of brain imaging) has included sulcal depth and brain connectivity as additional features in the PBM optimization. While promising, such approaches are specific to a particular data set, anatomy, and application. Geodesic distances to user identified landmarks on individual shapes were also used to guide the PBM optimization for complex shapes. However, this predefined mapping requires a careful selection of hand-crafted features and their success heavily relies on the expertise (and time) required to find such features consistently.
Here we propose an automated feature-learning approach for establishing dense correspondence models to alleviate the need for engineered shape descriptors or guiding landmarks. The idea is motivated by recent advances in computer vision and computer graphics that use deep convolutional neural networks (CNNs) to learn shape features for establishing pairwise correspondences between shapes. In leveraging the feature-based PBM, we effectively deep learn features to establish shape correspondence using direct surface geometry information rather than relying on predefined surface descriptors. We incorporate the deep learned features in the PBM optimization to automatically find a useful set of dense correspondences across complex shape ensembles. Therefore, we relax the Gaussian assumption of the current PBM approach while minimizing the required domain expertise by learning shape-specific features in a fully automated fashion.
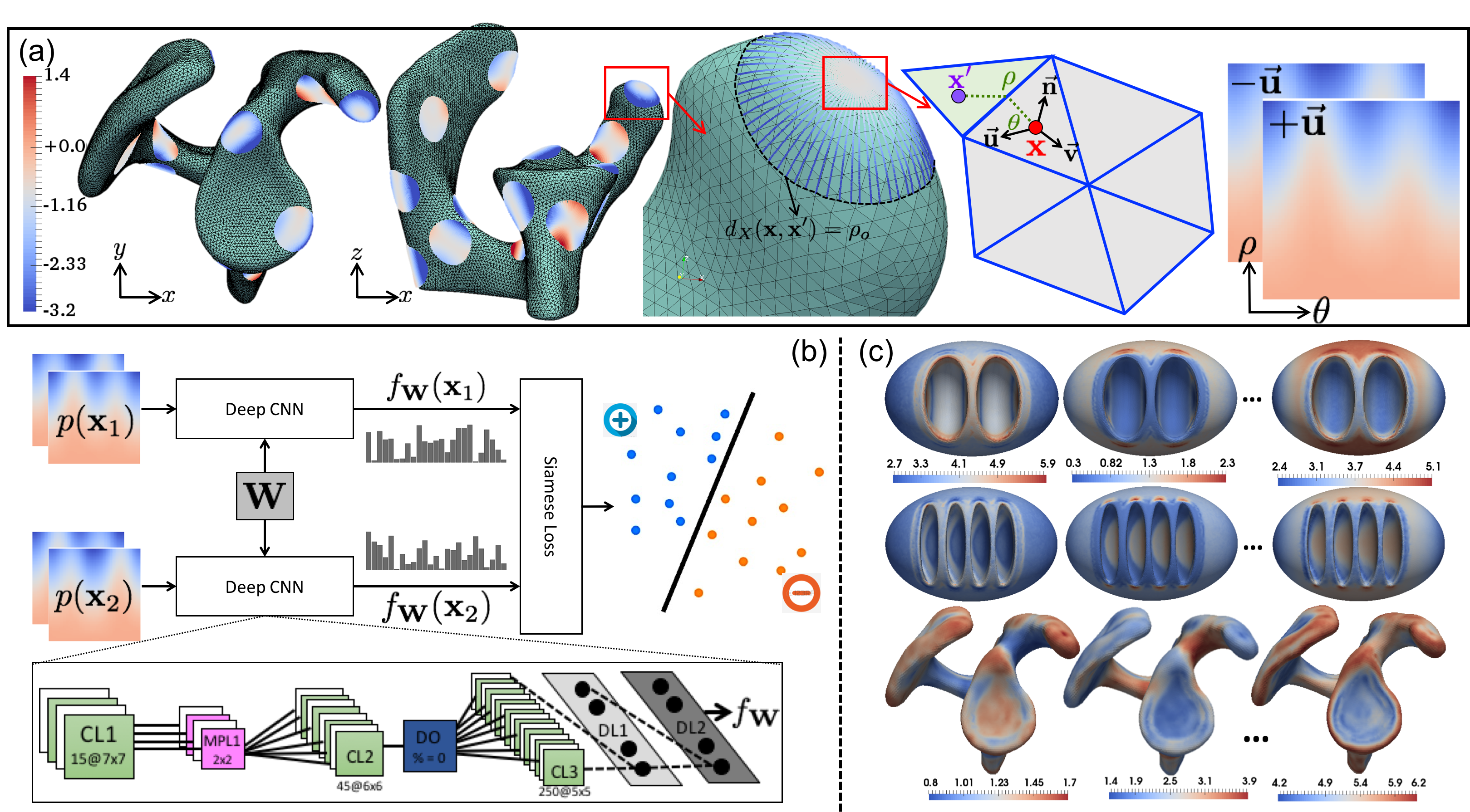
Proposed automated feature learning: (a) Illustration of geodesic patch extraction at a surface point. (b) The Siamese network architecture used to learn correspondence features. (c) Sample feature maps extracted using trained networks on synthetic and scapula shapes.
Related publications:
Praful Agrawal, Ross T. Whitaker, Shireen Y. Elhabian. Learning Deep Features for Automated Placement of Correspondence Points on Ensembles of Complex Shapes. 20th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2017.
Estimating shapes directly from images
Joint work with: Ross Whitaker, Ladislav Kavan, and Joshua Cates
Segmenting subject-specific anatomies is imperative to building statistical shape models (SSM); a time-consuming, expert-driven, expensive, irreproducible, and error-prone task. Some of these limitations can be addressed by (semi-) automated image segmentation approaches, but a robust, accurate, fully automated, and high-performance solution for different types of anatomies remains a challenge. To facilitate subsequent statistical analysis, segmentation is typically followed by a processing pipeline of shape registration and dense placement of corresponding landmarks, which entails significant parameter tuning, and/or quality control by the users. Hence, the significant time and effort required to prepare image data for SSM is a major barrier to making shape modeling a robust tool for on-demand clinical diagnostics, especially when dealing with large cohorts.
Therefore, in this work we address the problem of generating a rich set of shape descriptors in the form of PCA loadings on a shape space and an associated set of dense (i.e., thousands) of correspondence points. The goal is to design a system that bypasses the typical pipeline of segmenting and/or registering images/shapes and the associated optimization (and associated parameter tuning), and instead produces shape information directly from images via a deep (convolutional) neural network. This shape information can then be used to study pathologies, perform diagnoses, and/or visualize or study properties of populations or individuals.
DeepSSM is a deep learning approach that uses a convolutional neural network (CNN) to extract a low-dimensional shape representation directly from 3D images, requiring virtually no parameter tuning or user assistance. To overcome the challenge of the limited availability of training images with dense correspondences, we present a novel data augmentation procedure that uses existing correspondences on a relatively small set of processed images with shape statistics to create plausible training samples with known shape parameters. In this way, we leverage the limited CT/MRI scans into thousands of images needed to train a deep neural net.
We validated DeepSSM for three different applications pertaining to modeling pediatric cranial CT for characterization of metopic craniosynostosis, femur CT scans identifying morphologic deformities of the hip due to femoroacetabular impingement, and left atrium MRI scans for atrial fibrillation recurrence prediction.
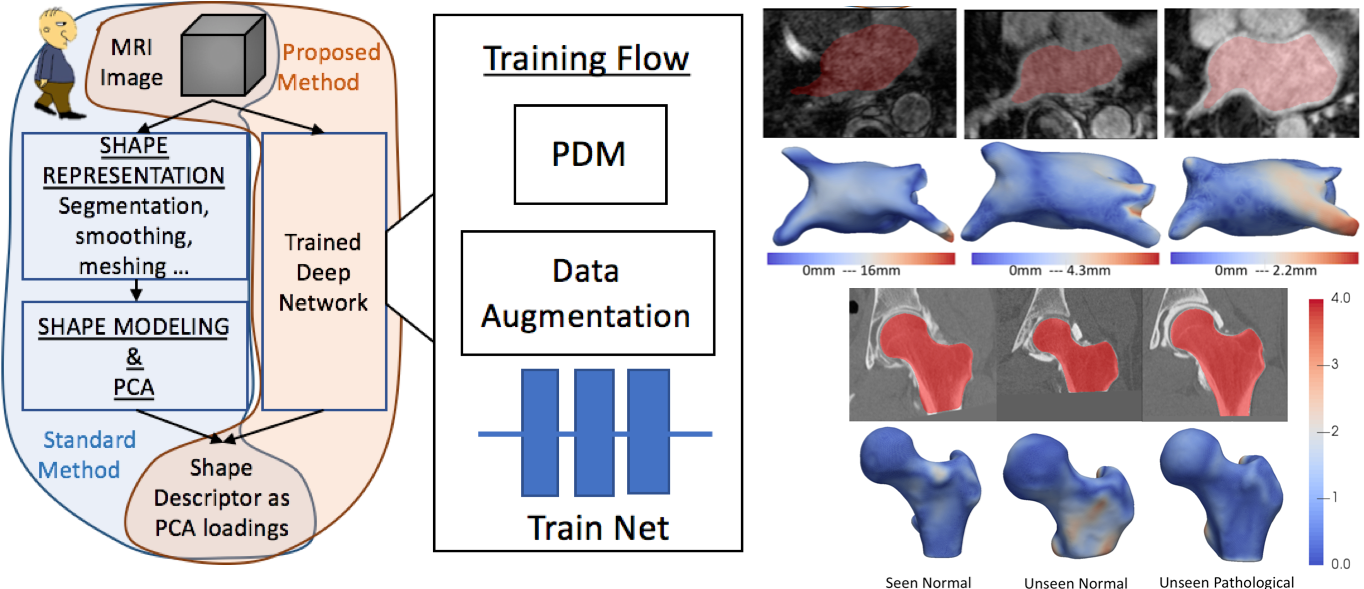
Left: the workflow of model training, it also shows the usage of the trained network for new images and is compared to the standard state-of-the-art workflow. Right: sample results on left atrium and femur anatomies.
Related publications:
Riddhish Bhalodia, Anupama Goparaju, Alan Morris, Evgueni Kholmovski, Nassir Marrouche, Joshua Cates, Ross Whitaker, and Shireen Y. Elhabian. Deep Learning for End-to-End Atrial Fibrillation Recurrence Estimation. Computing in Cardiology (CinC), 2018.
Riddhish Bhalodia, Shireen Y. Elhabian, Ladislav Kavan, and Ross T. Whitaker. DeepSSM: A Deep Learning Framework for Statistical Shape Modeling from Raw Images. ShapeMI-MICCAI 2018: Workshop on Shape in Medical Imaging, 2018.
ShapeWorks for modeling cranial nerves
Joint work with: Michel Audette and Ross Whitaker
We propose a segmentation methodology for brainstem cranial nerves using statistical shape model (SSM)-based deformable 3D contours from T2 MR images. To this regard, we modified ShapeWorks, the open source implementation of the particle-based shape modeling, to accommodate 3D contours. High-resolution T2 MR images are segmented for nerve centerline using a 1-Simplex discrete deformable 3D contour model. These segmented centerlines comprise training datasets for the shape model. Point correspondence for the training dataset is performed using an entropy-based energy minimization framework applied to particles located on the centerline curve. Shape statistics are incorporated into the 1-Simplex model by introducing a shape-based internal force, making the deformation stable against low resolution and image artifacts. A probabilistic digital atlas of the ten brainstem-attached cranial nerve pairs is constructed by incorporating a statistical shape model with the 1-Simplex deformable contour. The integration of shape information as a priori knowledge results in robust and accurate centerline segmentations from even low-resolution MRI data, which is essential in neurosurgical planning and simulations for accurate and robust 3D patient-specific models of critical tissues including cranial nerves.
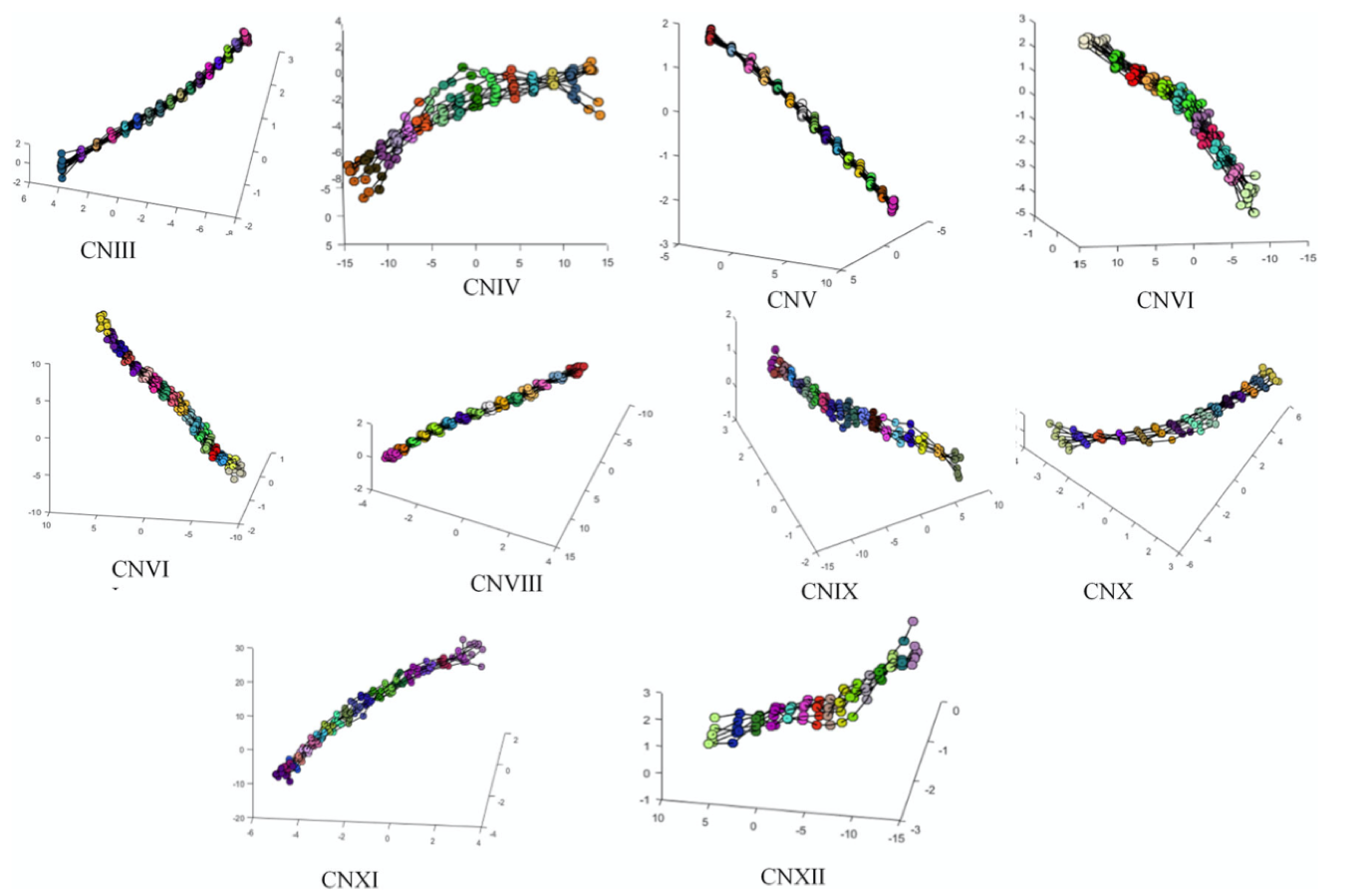
Correspondence models for cranial nerves III–XII
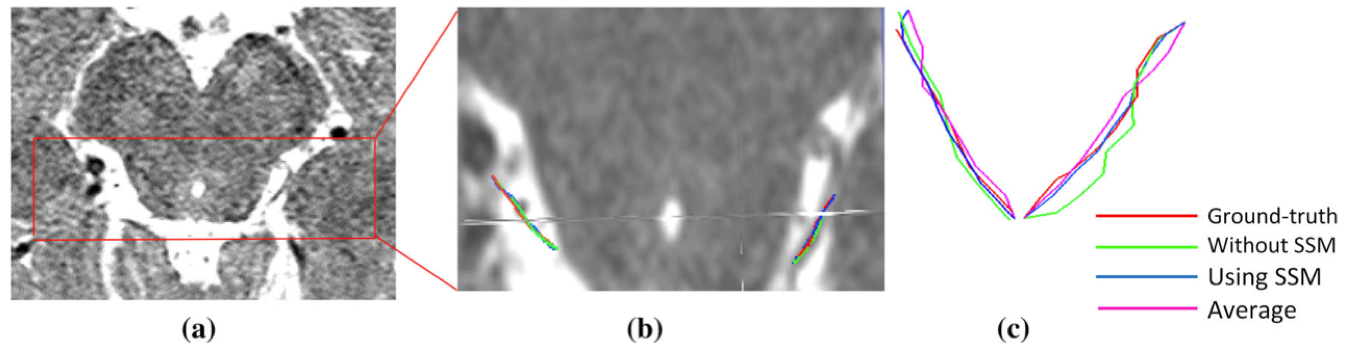
An example segmentation of CNIV from a partially visible dataset. (a) Cropped volume containing the nerve area; (b) magnification of the area bounded by red box in (a), along with the extracted centerlines; (c) left and right side of the nerve.
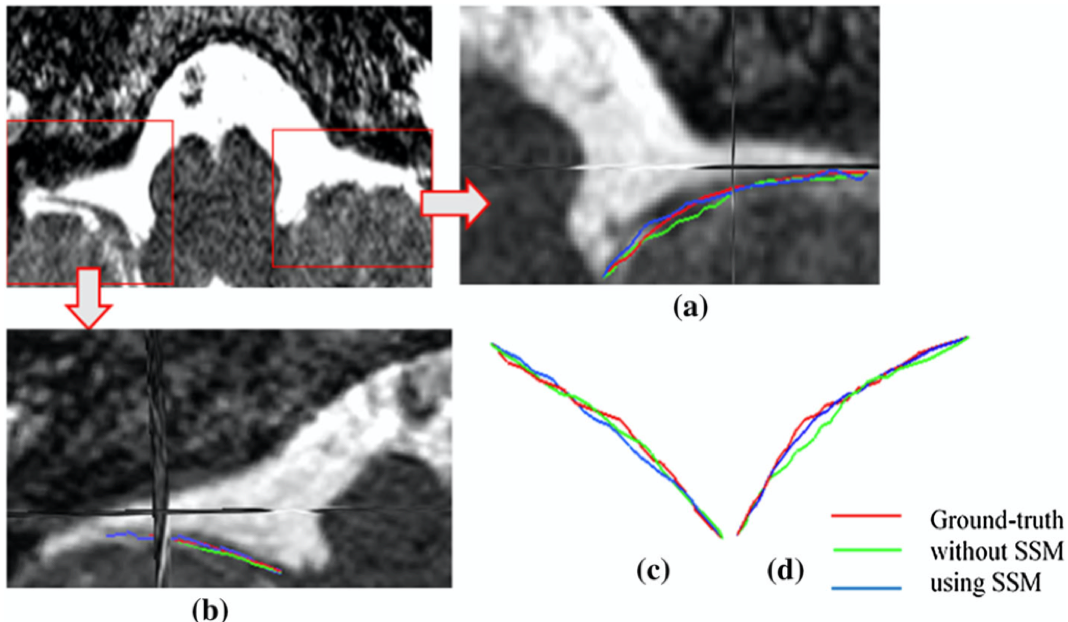
An example segmentation of CNX from a partially visible dataset. (a) Magnification of right side of the nerve which has partial volume effect; (b) magnification of left side of the nerve; (c,d) extracted centerlines.
Related publications:
Sultana Sharmin, Praful Agrawal, Shireen Y. Elhabian, Ross Whitaker, Jason E. Blatt, Benjamin Gilles, Justin Cetas, Tanweer Rashid, Michel A. Audette. Medial Axis Segmentation of Cranial Nerves Using Shape Statistics-aware Discrete Deformable Models. International Journal of Computer Assisted Radiology and Surgery, pp. 1-13, 2019.
Sultana, Sharmin, Praful Agrawal, Shireen Y. Elhabian, Ross T. Whitaker, Tanweer Rashid, Jason E. Blatt, Justin S. Cetas, Michel A. Audette. Towards a Statistical Shape-Aware Deformable Contour Model for Cranial Nerve Identification. In Workshop on Clinical Image-Based Procedures, Springer International Publishing, pp. 68-76, 2016.
Copyright © 2022 Shireen Y. Elhabian. All rights reserved.