 |
Here we describe the mechanics of accessing and manipulating files using the CVRTI graphicsio library. This section is most relevant to programmers; users of programs written for graphicsio files can happily skip this drivel entirely.
A library of functions and subroutines provide controlled access to CVRTI graphicsio time signal and geometry files. This approach serves to both relieve the user of some programming effort and also to allow some self-protection so that programming errors do not result in damage to file integrity or--worst of all--loss of information. Layered on top of the graphicsio library are a number of higher level utility functions, and self-contained tool programs, both of which facilitate developing and testing programs.
The design strategy for the routines that make up the graphicsio library was to first develop the core modules, internal data structures, and conversion routines, and then construct user-accessible routines on this base. To support a range of computer languages, we used standard ANSI C to create the data file routines, then wrote wrapper functions in Fortran. The Fortran routines do nothing more than perform any necessary data type conversions, call the underlying C functions, and then return the appropriate values. This way, code maintenance and development can focus on the C functions while still providing (almost) complete support for Fortran users. For reasons of compatibility, we restricted the Fortran to ``standard'' F77, which for practical purposes meant avoiding the more esoteric VMS extensions such as data structures as well as any of the Fortran90 features. To date, we have ported the library to Unix variants from Silicon Graphics, the IBM AIX, Sun OS, Linux, and Solaris with fairly minor alterations in the source code.
The exceptions to Fortran support for library capabilities was in a set of functions that simplify the editing or re-writing of data files and for the fiducial information in the tsdfc files. We may address the latter shortcoming, but then again, we may not...
Geometry files appear to the user fundamentally different from the tsdf files, simply because of the contents. Internally, however, the structure of both file types is virtually identical and many of the same data structures and internal functions are present in both. It is perhaps most accurate to view both the tsdf and the geometry files as subsets of a more general file structure. The essential elements of the file structure abstraction then become the main header, which contains enough information to describe the size and type of the blocks to follow, and the block themselves, which again must contain enough self-description to identify their character and content. The logical data structure for such a hierarchy is the linked list in which each block contains a pointer to the location of the start of the next block.
There are a number of conventions we have imposed on the users of the graphicsio library:
To simplify the manipulation of existing tsdf files, we developed a set of ``rewrite'' functions, based on a queue of edits or additions. Each time the user makes a change to the tsdf file, the request to change is queued, together with enough information to carry out the changes. A call to a flushing function initiates processing of the queue, which reconciles against the contents of the original file so that the new version contains all the contents of the original, plus any changes requested in the queue. Each entry of the queue consists of a code that reveals which file parameter is to be altered and a pointer to the new values for this parameter. These pointers can be simple pointers to data values or buffers but also pointers to the names of functions that, when activated, create the new values. It is this ability to point to the name of a function that prohibited Fortran77 support of re-writes.
We begin the detailed description of the programming steps required for the graphicsio files with a summary of steps required to write and read each type of file. Details of each step either follow here or are available in the list of calling functions and their arguments in Section 5.
The steps required to open a new geometry file and load it with data are as follows:
To read the contents of the file, the process is as follows:
For a sample of a function that reads a geometry file, see Appendix A.1.
The graphicsio library has an overall structure for handling tsdf files that is very similar to that for geometry files. Here, there is only one block, the time series, and a series of read/write (get/set) functions that manipulate specific elements of the time series.
The steps for creating a time series file are as follows:
To read the contents of the file, the process is as follows:
The block of time series signals that the graphicsio routine gettimeseries returns is multiplexed in one of two ways.
enum DataFormat {FORMAT_MUXDATA = 1, FORMAT_SCALARS, FORMAT_CVRTIRAW,
FORMAT_CVRTIPAK};
and the value for any tsdf file is returned by gettimeseriesformat.
The calling program is responsible for unpacking recovered data blocks or packing them for subsequent storage in a new tsdf file.
When the time signals in the tsdf data are external, the ``actual'' data files (.acq, .raw, or .pak file) are usually stored in other directories from the one containing the tsdf file. The default location for these files has traditionally been the CVRTI Vax, which is available via NFS as /vax/mapping/data for our systems. To define another path to the directory containing .acq, .pak, or .raw files, use the settimeseriesdatapath function before getting the time series data.
Some programs written at the CVRTI use an environment variable called MAP3D_DATAPATH that contains the desired path for the .pak and .raw files.
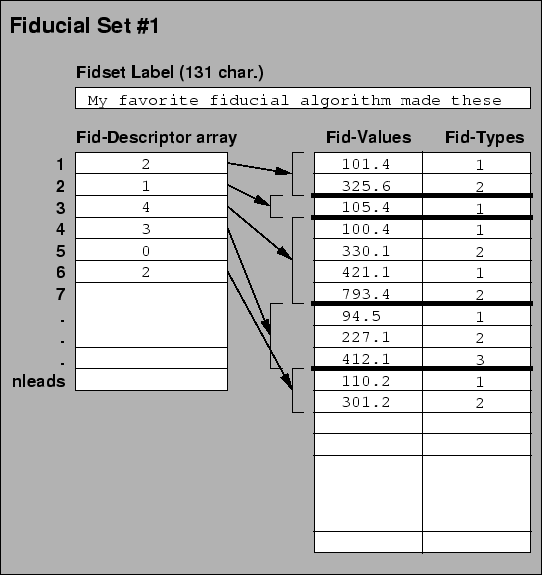
The challenge in describing fiducial information is to permit a large range of flexible options for defining, detecting, and storing fiducials within the file structure in a clear manner. To this end we have developed the structure shown in Figure 3.1
Note the following features of this organization:
To simplify and coordinate assignment and management of fiducial types, we have created a library that gives access to a fiducial type database. This library is described in detail at www.cvrti.utah.edu/~dustman/fi. Use of this library will ensure that the most current links between fiducial types are their text names is available to your program.
Section 2.1.3 described the organization of the container files, i.e., the files that are containers to a series of tsdf files. The container files also contain information extracted from the time series, eg., temporal fiducials determined on a channel-by-channel (or lead-by-lead) basis. Certainly the most frequent example is the activation time, which we routinely compute.
At present, there are no routines within graphicsiofor handling fiducials in tsdfc files. Examples of programs and libraries that do provide support for tsdfc files include Everett, a program by Ted Dustman for initial processing of mapping data, Matmap, a set of MATLAB uilities by Jeroen Stinstra with a similar functionality, and tsdflib (as yet undocumented), a library created by Ted Dustman, Rob MacLeod, Jenny Simpons, and Jeroen Stinstra that provides C-language access to container files.
If you are planning to program with graphicsio, there are two ways to go. If all that is required is access to the functions, then linking to libgraphicsio.so is the most sensible approach. Recent versions of the library with both C and Fortran entry points are always available in /usr/local/lib (and /usr/local/lib64 if you need a 64-bit version).
If you want know more or actually add functionality to graphicsio, you should coordinate with Ted and make use of the CVS repository that we have for the graphicsio source code. For details on this, see www.cvrti.utah.edu/~dustman/cvs-graphicsio
With the ubiquity of Matlab has come the need to provide an interface to graphicsio files from Matlab, which we have accomplished in two ways. The first was a stand-alone program that converts tsdf files into Matlab ``mat'' files. This approach has its limitation and so we have largely abandoned it for the more elegant method of direct access through Matlab.
To add functionality to Matlab, one creates ``mex'' files that contain some Matlab and some C code and the result is new commands that appear transparent to the Matlab user. This is all explained in the Matlab manuals, but the main result of all this are three new Matlab commands:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
To make sure you have these command available to you, add the following to
the path variable in Matlab:
/usr/local/matlab/datamex