 |
In this section we describe the logical structure of the graphicsio file system. The aim is not to provide all implementational or usage details (that will come in Section 3) but to offer all users of the graphicsio system some insight into the design and contents of the files. Hence, the following should be useful not only for programmers, but especially for users of programs that access graphicsio files.
The external view of the CVRTI graphicsio file system is of two different file types, one that holds sampled time signals and the other that contains descriptions of geometry. Internally, we have implemented these as two facets of the same basic structure in order to share many internal data structures and access routines and thus reduce code development effort. User or programmer-level access to both files occurs largely through type-specific library functions, hence it seems suitable to maintain the logical separation of file types in the description that follows.
The need for portability across operating systems required us to adopt a single storage format with internal conversion routines to take care of the byte-order swapping and difference in floating point storage that different computers require. Because we had more previous experience with VMS than any other system, we chose this as the basis for determining internal data representation--the graphicsio files are ``native'' to VMS. Access from outside VMS is supplemented by on-the-fly conversion routines that deal with all operating system differences. As a result the files themselves may be copied from system to system without regard to local data presentation standards or operating system. At this point, we have implemented the graphicsio file routines for VMS and a variety of Unix versions including SGI Irix, IBM AIX, Linux, and SunOS and Solaris for Sun workstations.
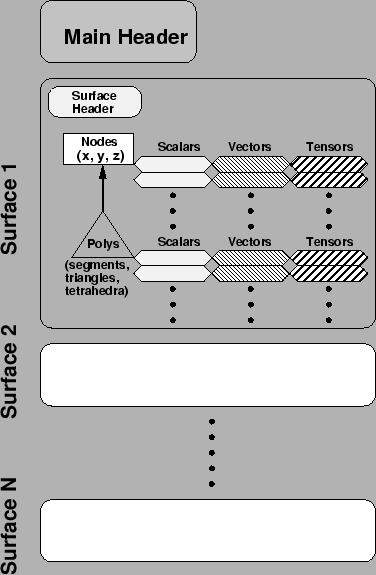
The purpose of the geometry files is to store the locations of sets of points in three-dimensional space, any connectivities that bind them into a mesh, and any scalar, vector, or tensor quantities that are associated with the points or polygons of the mesh. For the structure of the geometry files, we have chosen to organize these points into ``metasurfaces''. More accurately, a metasurface combines nodes and connectivity information; the connectivity can be in the form of line segments, triangles, or tetrahedra (with other polygons possible should the need arise). To each point or polygon of each surface we can associate additional quantities--scalars, vectors, and tensors are currently supported. Physical examples would be electrical conductivity--either scalar or tensorial--at a point; a normal vector at a point or for a planar polygon; or a value which defines the tissue type or anatomical context of a point or element. What we do not associate with geometry are time signals; these live in a separate file type described in Section 2.1.3.
The logical structure of the CVRTI geometry file format is shown in Figure 2.1. The hierarchal structure of the geometry file is a tree with a single header at the top containing file type, text description, number of surfaces, etc.,. Below the main header are the metasurfaces, each of which contains points, then optionally one or more sets of connectivities based on polygons. Below each set of points or polygons are the associated sets of scalars, vectors, or tensors. Each geometry file contains some subset of all the possible elements shown in the figure, however, at a minimum a geometry file must contain just one point; everything else is optional. There is no limit (other than that dictated by 4-byte long-integer representations in the computer) to the number of nodes, polygons, or associated values a surface may possess, nor in the number of surfaces in a single file. The different metasurfaces of a file typically represent physically different sets of points, for example, one surface of measurement electrodes on the heart surface and a second one from the body surface, or temperature sensors divided into one surface at ground level, one at 5,000 feet and a third at 10,000 feet in the atmosphere.
There is often confusion with the meaning of the terms ``leads'', and ``channels'' and their association with nodes in the geometry. As we will see in the next section, time signals are organized into channels of data, each representing the stream of numbers that passed through a specific channel of an acquisition system. Leads also refer to time signals, but usually have meaning in the specific context of the sensor location (and not necessarily the input amplifier of the acquisition system). For example, a lead might refer to the standard ECG leads on the torso surface called V1- V6 and we often need to know what the corresponding channels in the data stream were, and to which nodes of the torso geometry they correspond. The sensor labeled lead- V1 may be located at node J and the time signal from this sensor may be acquired through amplifier channel K. If J = K then the link between node and channel can be implicit and the numbering schemes are superimposed. If, as is more often the case, the numbering schemes differ, then some means of linking them is necessary. The leads and channels arrays (and associated files) take care of this linking.
The figure 2.2 shows an example of lead, channels, and channel-links information for a hypothetical case. See the figure caption for details.
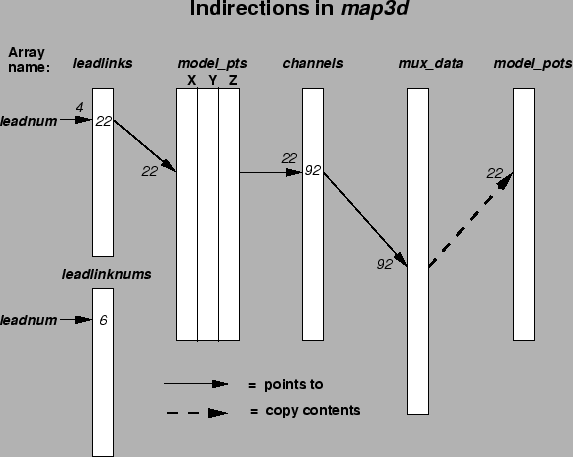 |
Leadlinks, channels, and channellinks files are separate entities outside of the graphicsio file system and are described in detail in the map3d manual. Channels information can also reside in a graphicsio geometry file, as an associated scalar value to each node. Because channel links are usually stable for an entire experiment or set of simulations, we have chosen the geometry file rather than the time signal file as the logical place for this information. At present, there is no place in the graphicsio files for leadlinks or channellinks information.
Time signal files in the graphicsio format have similar features to the geometry files. Just as the surface is the basic unit of the geometry file, time signal files are based on contiguous blocks of scalar values, which we refer to as ``time series''. Each time series is organized into individual data channels which we require be spaced at regular intervals in time. Viewed one way, each time series is a set of spatially organized channels of regularly sampled time signals of a scalar quantity; viewed orthogonally, each time series is a sequence of distributions of a scalar quantity sampled synchronously over all channels at a regular time intervals. Expressing the latter view in the language of electrocardiographic mapping, each distribution is a single ``map'' so that we have a sequence of maps in each time series.
One difference between geometry and time series files is that the latter is really a hierarchy of files, which allows flexible grouping of time series. At the lowest level are the time series data files (tsdf), which contain a single time series. The time series data file container (tsdfc) files hold pointers to the individual tsdf files and parameters extracted from these times series. Data file container (dfc) files provide further levels of grouping, if desired. Below we describe this hierarchy in more detail.
At the lowest level in the time series file hierarchy are the ``time series data'' or ``tsdf'' files with the file extension .tsdf. Each tsdf file contains a single time series, that is, a single, continuous recording, and all the associated information necessary to unpack and interpret these signals. 2.1 Below is a table of the parameters that are stored in the tsdf file.
| ||||||||||||||||||||
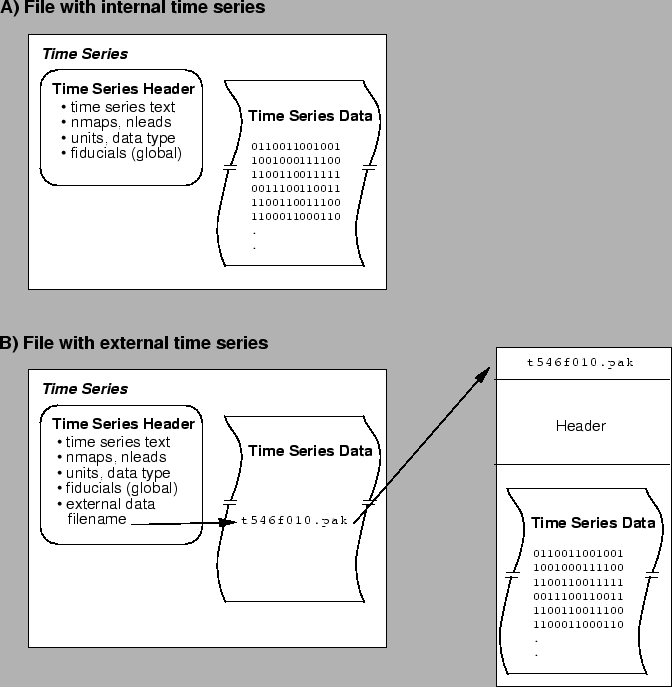
The time series data itself can be stored in two forms within the tsdf file. Figure 2.3 shows both forms, which we call ``internal'' and ``external'' storage. In the case of internal storage, shown in the upper panel of the figures, the time series data values are part of the tsdf file. In external mode, shown in the lower panel, the tsdf file contains a pointer (a file name) to another file that contains the actual time series. The external file can be in one of three CVRTI formats:
 |
The internal ordering of the scalar times signals in a tsdf file is described by the value of the format parameter. The most common format reflects ordering by distribution or map, i.e., the first value is channel #1, time instant #1; then channel #2, time instant #1; channel #3, time instant #1, etc.,, which we refer to as ``multiplexed ordering''. The complement is ordering by channel: channel #1, time instant #1; then channel #1, time instant #2; channel #1, time instant #3, etc.,.
The graphicsio routines also permit access to either the entire time series at once, or any piece of a time series, thus permitting a better trade-off between time series length and available program memory. Individual data values are stored as single precision, floating point numbers, (4 bytes per value). This level of resolution is more than adequate for most measured data, which is typically limited to 12-16 bits. To mesh more easily with programs that use other data storage conventions, there are graphicsio routines that return time series values in other formats (eg., double precision, integer.)
To gather a number of tsdf files into a single unit, we have defined the ``time series data file container'', or ``tsdfc'', files. Each tsdfc file contains no actual time series values, i.e., no signals, but pointers to the tsdf files that contain the time series data. So each tsdfc file is just what it says, a container for one more more tsdf files. In addition, the tsdfc file can contain parameters that are derived or extracted from the time series. The most common examples of this at the moment are time fiducial parameters, eg., activation and recovery times. The idea is generalizable and extensible, however, so that we can define any number of parameter types and then include them in the tsdfc format.
Internally, the tsdfc file is actually a Gnu Database Manager (gdbm) file (see www.delorie.com/gnu/docs/ for more details), which simplifies management of the file contents and builds on existing standard and libraries (this is Unix, after all...). We have written, and are in the process of writing more, layered libraries to handle the tsdfc files that will become part of the graphicsio library. See Ted Dustman's EIO library for an example as well as his Everett program.
Each record in the tsdfc file contains a key value, which is the name of the tsdf file to which it refers. Together with this key are zero or more sets of parameter sets. Again, there are no time signals in the tsdfc file, just pointers to the files that contain those signals.
The hierarchy of time series files can continue, in principle infinitely, with containers that contain lower level files. At this point, we have only one additional file type, the ``data file container'' or dfc file, which can contain multiple tsdfc files and also multiple dfc files, hence it is a recursive design. An example of such groupings would be a single experiment based on multiple subgroups of files, in which the data recorded before an intervention might be in one tsdfc file and those after the intervention in a second tsdfc file. The combination of two or more such groups could define a dfc file for the entire experiment.
As described in Section 2.1.2 above, the relationship between individual time signals from a time series file, the ``channels'', and locations stored in the geometry files, the ``nodes'', can be coded in the geometry file as a set of scalar values associated with the nodes. With this flexible mapping between data and geometry, time series files may contain signals from any number of surfaces or even multiple geometries. Either the associated geometry file or a separate channels file contains the information required to extract the desired channels and associate them with the proper node locations.
Channel attributes2.2 describe that state of each data channel. The current list of attributes coded in graphicsio includes:
| Valid channel attributes | |
| Good | no special attribute |
| Bad | the channel has technical problems that make its use questionable |
| Blank | the channel has no data at all |
| Interp | the channel contain interpolated data |
The list of channel attributes will continue to grow over time and the header file gi_graphicsio.h will always contain the latest list.
Each tsdfc file can contain zero or more sets of parameters that are
derived from the associated time series data and the best developed example
of a class of parameters are the fiducials. Fiducials are simply markers
of important events in the time course of the signals, for example onset of
the QRS, peak and end (offset) of the T wave, activation time, recovery
time, etc.,. They can be described in terms of frame numbers relative to
the samples in the time series, or as real values in time relative to some
arbitrary zero (which is a fiducial as well). In the graphicsio file
system, fiducials can be global in scope, effective for the entire
set of channels, or local to each channel. A graphicsio tsdf
file should no longer contain any fiducials-these reside in the tsdfc
file. For more information on fiducials, see Section 3.3.4,
below.