Kannan U V's
Portfolio et al.
Research
For the past 2 years, I have been developing algorithms using image processing and machine learning techniques for Collaborative Research in Computational Neuroscience (CRCNS) project. I have been working on machine learning based image segmentation, developing algorithms to segment cellular structures like cell membranes and synapses in neural tissues (Rabbit retina & C. elegans worm) scanned using Electron microscope.
Thesis: My master's thesis is on "Automatic markup of neural cell membranes using boosted decision stumps".
Publications
Kannan U. V., M. Kim, D. Gerszewski, M. Hall, “Assembling large mosaics of electron microscope images using GPUs"; Symposium on Application Accelerators in High Performance Computing (SAAHPC’ 09)[Accepted]
Kannan U. V., A. Paiva, E. Jurrus, T. Tasdizen, "Automatic markup of neural cell membranes using boosted decision stumps", IEEE International symposium on biomedical engineering (ISBI 2009) [Accepted]
J. Anderson, B. Jones, J. Yang, M. Shaw, C. Watt, P. Koshevoy, J. Spaltenstein, E. Jurrus, Kannan U. V., R. Whitaker, D. Mastronarde, T. Tasdizen, R. Marc, “Ultra structural mapping of neural circuitry: A computational framework” ", IEEE International symposium on biomedical engineering (ISBI 2009) [Accepted]
J. Anderson, B. Jones, J. Yang, M. Shaw, C. Watt, P. Koshevoy, J. Spaltenstein, E. Jurrus, Kannan U. V., R. Whitaker, D. Mastronarde, T. Tasdizen, R. Marc, “A Computational Framework for Ultrastructural Mapping of Neural Circuitry”, PLoS Biology
Software
ir-assemble-CUDA
About: ir-assemble is a tool in the neural circuit reconstruction pipeline used assemble mosaic of electron microscope images. I was involved in accelerating the application by rewriting the ITK based multithreaded application in CUDA and achieving 12x acceleration.
Synapse Viewer
About: This viewer is built based on Qt & C++. This is a very primitive viewer just to open up the image and see the synapse markup. Using this software one can scale and scroll through the image data. One can compare multiple markup sets (like the ground truth and machine learning algorithm predicted synapse structures simultaneously).
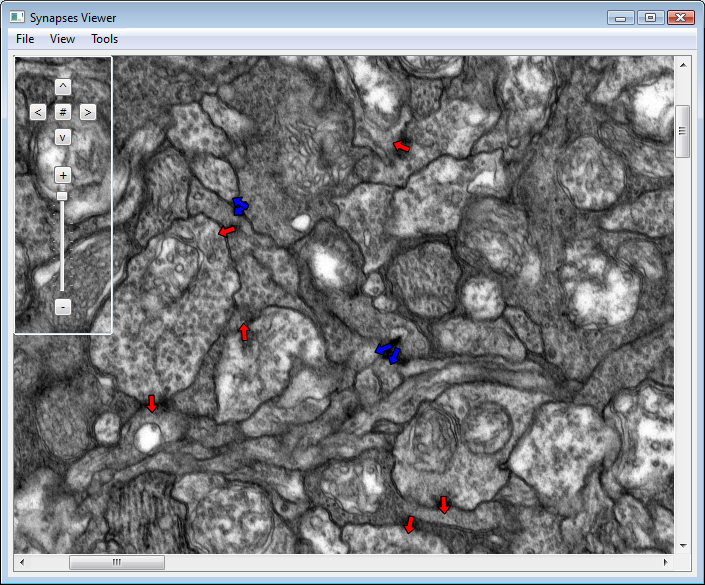
Checking out Code: The code is available in the SVN repository for CRCNS. The repository can be accessed from https://gforge.sci.utah.edu/
The code is available in Code\SynapseViewer of the CRCNS trunk
Building the Code in Linux & MinGW: The Qt program has the *.pro file. So the following commands would compile the project.
1) qmake
2) make
Building Code in MS Visual Studio 2008 Pro:1) You need to install QT open source building it from source.
You need to run the batch file C:\Program Files\Microsoft Visual Studio 9.0\VC\vcvarsall.bat. Set environment variable QMAKESPEC to win32-msvc2008 by "set QMAKESPEC=win32-msvc2008". Then run the configure in the source.
2) Generate make files. qmake
3) Generate an MS VC project & Solution by qmake -t vcapp
4) Then you can compile SynapseViewer from Visual Studio
