 |
Fluorescence Microscopy Designed especially for neurobiologists, FluoRender is an interactive tool for multi-channel fluorescence microscopy data visualization and analysis. |
Overview
Features:
- Clustering algorithms. Three clustering algorithms for segmentation are added: expectation-maximization on Gaussian mixture, DBSCAN, and k-means. Users can use these methods in the "Component" window.
- Improved tracking algorithms. Tracking accuracy has been improved. We improved the algorithm for generating the track map and incorporated clustering algorithms to automatically segment during tracking. Users can also adjust a series of parameters in the "Tracking" window to fine tune the tracking.
- A new 4D script for tracking sparse particles. The 4D script allows users to track selected features in a time sequence. No initial segmentation is required for it to work. It can be used to track sparse and small features conveniently.
- A density setting for component generation. The density setting has been added in the basic component generation. Its concept is based on the clustering algorithm DBSCAN. However, its implementation is based on the synthetic brainbows algorithm, which uses GPU to compute segmentation of dense data sets.
- Render view output enlargement. The option has been added when the render view is captured and saved as an image. Users can set an output image size larger than the render view size.
- 4D script list. The list of built-in 4D scripts has been added to the "Record/Export" panel. Users can easily select and switch 4D scripts without browsing to the actual files.
The MIT License Copyright (c) 2012 Scientific Computing and Imaging Institute, University of Utah. License for the specific language governing rights and limitations under Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
If you use FluoRender in work that leads to published research, we humbly ask that you add the following to the 'Acknowledgments' section of your paper:
"This work was made possible in part by software funded by the NIH: FluoRender: Visualization-Based and Interactive Analysis for Multichannel Microscopy Data, 1R01EB023947-01 and the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P41 GM103545 and R24 GM136986.”
Requirements
Basic system
- OS: Windows (64 bit) or Mac OS X
- CPU: Intel or AMD 64-bit processors
- Graphics card: Nvidia or AMD graphics cards
- System memory: 4GB
- Display resolution: 1280 x 1024
Example high performance system(s)
- OS: Windows 10 Pro
- Motherboard: Asus 270-WS
- CPU: Intel Core i7 7700K
- Graphics card: Nvidia Geforce GTX 1080 Ti, Quadro P6000 (10-bit support), or AMD Radeon RX Vega, FirePro WX9100 (10-bit support)
- System memory: 64 GB
- RAID: 2x Samsung NVMe SSD 960 PRO 1TB in RAID0
- Display: Dell UltraSharp UP2716D (10-bit support)
Note: Only matching professional-level graphics cards and displays can support 10-bit output. Refer to the FluoRender user's guide for setting 10-bit output.
FAQ
1. What computer hardware is good for running FluoRender?First of all, FluoRender is built to run on a personal computer with good graphics processing capabilities. Although most of today's personal computers can run FluoRender, including Windows desktops, laptops, Apple MacBooks, iMacs, and MacPros, it is best to purchase one with a gaming level or professional graphics card. Wikipedia provides detailed comparisons of graphics card from both Nvidia and AMD. Generally speaking, a graphics card with higher "Processing Power GFLOPs" is also better for FluoRender. We would recommend a top-of-the-line gaming graphics card over a professional graphics card. FluoRender can take advantage of some features of a professional graphics card, such as 10-bit output and better anti-aliasing. However, the difference is subtle in most situations. Second, strong CPU processing power is generally desired. We would recommend a latest CPU with higher clock frequency over more cores. Third, if you have large data sets to process, equip as much system memory as possible. For the year 2015, the consumer level computers can be equipped with as much as 64 GB of system memory, while a professional desktop can have 512 GB or more. Last, if you want to load data quickly or play back large time sequence smoothly, you need high speed access to hard drives. We would recommend a discrete RAID controller with abundant cache. Speed of hard drives is also important for handling large data sets. We would recommend 4 or more solid state drives configured in RAID0 mode.
2. Why can't I start FluoRender?
There are several possibilities for FluoRender to fail. First, check if the graphics driver is installed and updated to the latest version. Some old graphics card may not support OpenCL, which is required since version 2.15. If that is the case, you can download and try an earlier version of FluoRender. Since version 2.16, we require the graphics card to support at least OpenGL 3.3. If you have an old graphics card and want to use FluoRender on it, please replace your graphics card or download an old version of FluoRender. Second, make sure that your operating system supports 64-bit applications. Since version 2.15, we have dropped the support of 32-bit applications, which means FluoRender is 64-bit only. For Windows, you have to purchase the x64 version of the operating system; for Mac OSX, you have to update to the recent versions. Finally, FluoRender may not be properly installed, or some required modules have been accidentally uninstalled. Reinstalling FluoRender may solve the issue.
3. Why can't I load a file into FluoRender?
FluoRender can only load supported file formats, which include TIFF and some microscopy manufacturer specific formats. We write our own readers for these formats to achieve the best performance, especially for large data and time sequence data. So, we can only support "Open" and standard formats. If you have files of unsupported formats, you have to convert the format using a third-party tool. ImageJ can be a good choice in most situations.
4. Why can't I use some of the functions in FluoRender?
If you can use FluoRender for the basic visualizations of volume data, but not some advanced functions, such as paint brushes, it means your system does not support all OpenGL features. This can be that you are using a virtual machine, a remote client, or a system with an integrated graphics card with limited capabilities. You may want to upgrade your system to a standalone desktop computer with the latest graphics hardware to use all features in FluoRender.
5. Does FluoRender have a Linux version?
The official FluoRender releases only include a Windows version and a Mac OSX version, both 64-bit. If you want to run FluoRender on systems other than these, you can download the source code of FluoRender and build it on a different operating system. Additionally, you can send us emails and let us know your requirements.
6. How can I fix FluoRender crashes when rendering and computing take long time?
You may add a registry value, called "TdrLevel", on Windows. More information can be found here.
Sample Data
1. Zebrafish HeadConfocal dataset of 5 day old transgenic zebrafish larva, stained for anti-actin, anti-GFP (recognizing the isl1:gfp transgene), and ToPro3 (nuclear stain). Voxel size = 1.25 x 1.25 x 3.0 microns.
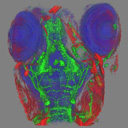 isl1actinCy3Top3_actin.tif [35MB]
isl1actinCy3Top3_actin.tif [35MB]
Support
Developers:
Yong Wan, Ph.D
This email address is being protected from spambots. You need JavaScript enabled to view it.
Scientific Computing and Imaging Institute, University of Utah
Advisors:
Chuck Hansen
Scientific Computing and Imaging Institute, University of Utah
Chi-Bin Chien (decesed)
Dept. Neurobiology & Anatomy, University of Utah
Alumni:
Brig Bagley, Research Software Engineer
This email address is being protected from spambots. You need JavaScript enabled to view it.
Hideo Otsuna, Ph.D
This email address is being protected from spambots. You need JavaScript enabled to view it.
Papers:
Hideo Otsuna, Yong Wan, Chi-Bin Chien and Charles Hansen. "Interactive Extraction of Neural Structures with User-Guided Morphological Diffusion", Publication No. US20140104273 A1. Application No. US 14/051,947.
Yong Wan, Hideo Otsuna, Kristen Kwan and Charles Hansen. "Real-Time Dense Nucleus Selection from Confocal Data", Eurographics Workshop on Visual Computing for Biology and Medicine, 2014.
Yong Wan*, Hideo Otsuna*, Chi-Bin Chien and Charles Hansen, "Interactive Extraction of Neural Structures with User-Guided Morphological Diffusion", in Proceedings of 2012 IEEE Symposium on Biological Data Visualization (BioVis), 2012.
Yong Wan*, A. Kelsey Lewis*, Mary Colasanto, Mark van Langeveld, Gabrielle Kardon and Charles Hansen, "A Practical Workflow for Making Anatomical Atlases in Biological Research", IEEE Computer Graphics and Applications, vol. 32, no. 5, 2012, pp. 50-60.
Yong Wan*, Hideo Otsuna*, Chi-Bin Chien and Charles Hansen, "FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research", in Proceedings of 2012 IEEE Pacific Visualization Symposium (PacificVis), 2012, pp. 201-208.
Yong Wan*, Hideo Otsuna*, Chi-Bin Chien and Charles Hansen, "An Interactive Visualization Tool for Multi-channel Confocal Microscopy Data in Neurobiology Research", IEEE Transactions on Visualization and Computer Graphics, vol. 15, no. 6, 2009, pp. 1489-1496.
Joining the FluoRender Users Mailing List
- Compose an email to This email address is being protected from spambots. You need JavaScript enabled to view it. with the following in the body of message: subscribe fluorender
- Nothing needed in the Subject line. Just send.
- You will receive a response from the list manager requesting confirmation. Just send back a reply message to confirm your subscription (no added text needed).
- Finally, you will receive a welcome message confirming that you have been added to the mailing list.
Unsubscribing
- Compose an email to This email address is being protected from spambots. You need JavaScript enabled to view it. with the following in the body of message: unsubscribe fluorender
- Nothing needed in the Subject line. Just send.
- You will receive a response from the list manager notifying you that the command has succeeded and you have been removed from the list.
