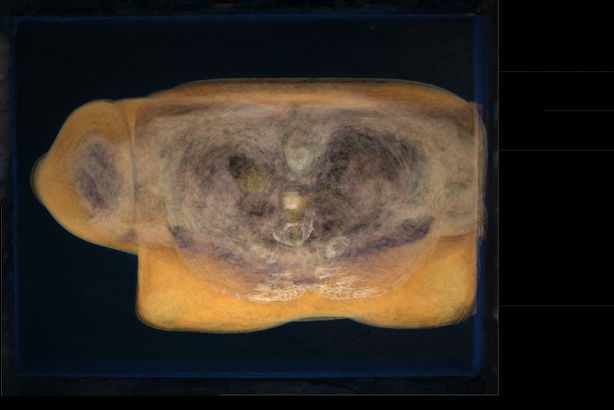
var-small.png
First, cropping. All the "Every 100th" data was downloaded into directory "every100th". The data was kept compressed, and NRRD headers were generated:
foreach SLC ( `echo *.raw.gz` )
set numraw = ${SLC:r}
set num = ${numraw:r}
echo $num.nhdr
unu make -h -i ./$SLC -t uchar -s 3 6144 4096 -c $numraw -e gz -o $num.nhdr
end
foreach NHDR ( `echo *.nhdr` )
echo ${NHDR:r}-small.ppm
unu resample -i $NHDR -s = x0.1 x0.1 -k cubic:0,0.5 -o ${NHDR:r}-small.ppm
end
unu join -i ????-small.ppm -a 3 \
| unu project -a 3 -m variance \
| unu gamma -g 2.5 | unu quantize -b 8 -o var-small.png

Based on this, minimum (X,Y) of (1550, 950) and maximum (4499, 3099) was chosen, and then tested:
foreach NHDR ( `echo *.nhdr` )
unu crop -min 0 1550 950 -max 2 4499 3099 -i $NHDR \
| unu resample -s = x0.2 x0.2 -k tent \
| unu gamma -g 1.4 -o ${NHDR:r}-test.ppm
end
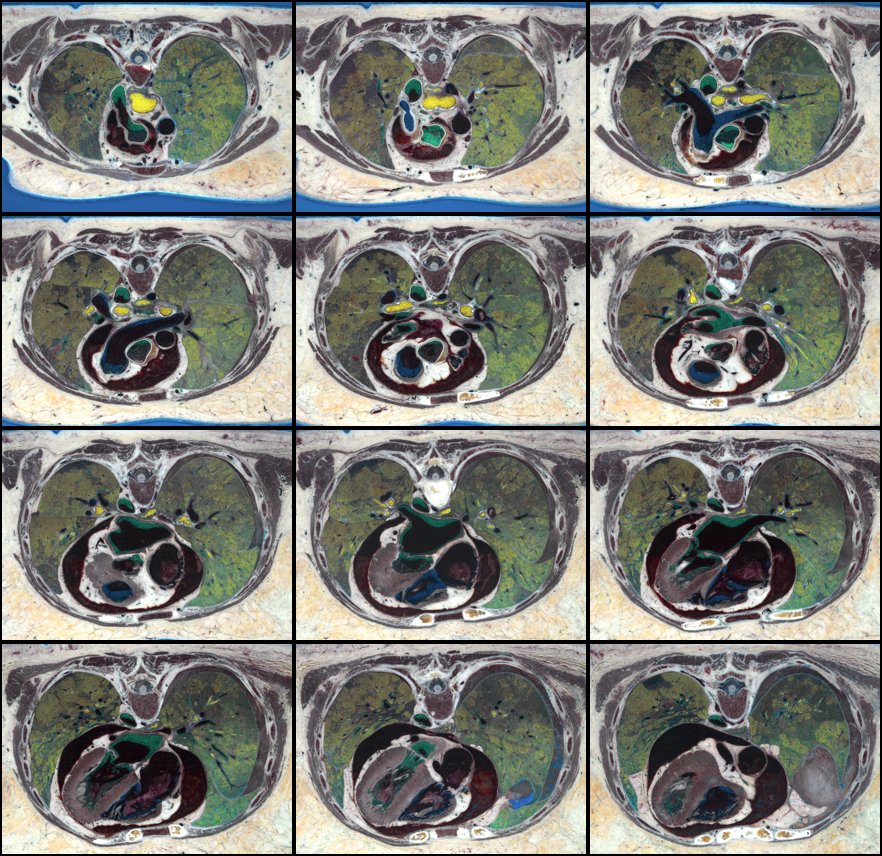
unu join -i *-test.ppm -a 3 \
| unu pad -min 0 -5 -5 0 -max 2 M+5 M+5 M -b pad -v 0 \
| unu reshape -s 3 600 440 5 6 \
| unu permute -p 0 1 3 2 4 \
| unu reshape -s 3 3000 2640 \
| unu resample -s 3 x0.25 x0.25 -o crop-test.png
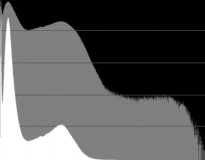
Now, quantization. I used the 16-bit slices to experiment with different parameters to adjust the values prior to quantization. First, NRRD headers were generated for all the 48bit slices, and then these were cropped according to the cropping parameters determined above, and then volumes of red, green, and blue values were generated in order to produce histograms of the color components.
foreach N ( 24 25 26 27 28 29 30 31 32 33 34 35 )
echo ${N}00.48.nhdr
unu make -h -i ./${N}00.48.raw.gz -t ushort -s 3 6144 4096 \
-e gz -en big -c ${N}00.48 -o ${N}00.48.nhdr
unu crop -min 0 1550 950 -max 2 4499 3099 -i ${N}00.48.nhdr \
-o ${N}00-crop.nrrd
unu resample -i ${N}00-crop.nrrd -s = x0.35 x0.35 -k tent \
-o ${N}00-small.nrrd
end
unu join -i ??00-small.nrrd -a 3 -o small.nrrd
unu dice -i small.nrrd -a 0 -o small-C
foreach C ( 0 1 2 )
echo small-C${C}-histo.png
unu histo -i small-C${C}.nrrd -min 160 -max 65540 -b 1024 \
| unu dhisto -h 800 -o small-C${C}-histo.png
unu resample -i small-C${C}-histo.png -k tent -s x0.2 x0.2 \
-o small-C${C}-histo-th.png
end
| 
| 
|
| small-C0-histo-th.png | small-C1-histo-th.png | small-C2-histo-th.png |
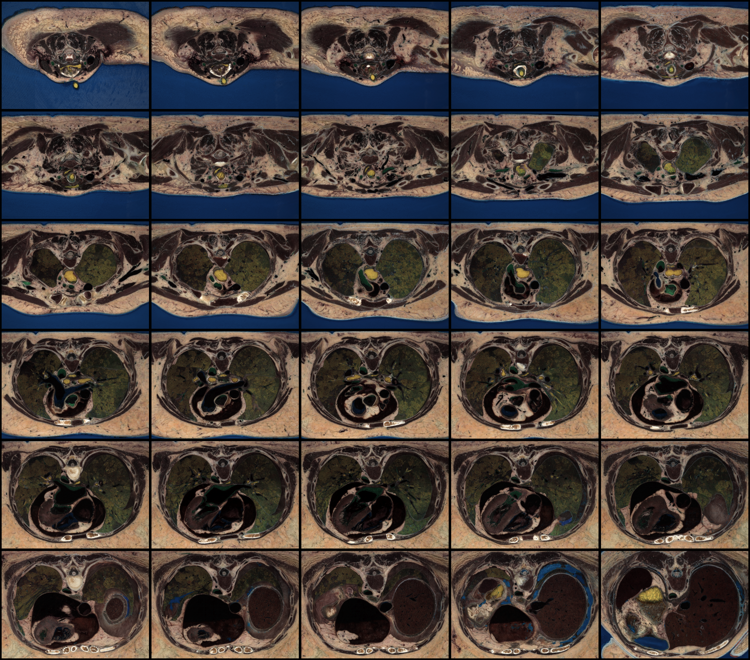
At this point I wanted to visualize the mapping used to generate the 8-bit images made available so far. In a seperate "inspect" directory:
foreach NUM ( 2400 3200 2900 3500 )
echo $NUM
unu jhisto -i ../48bit/$NUM.48.nhdr ../every100th/$NUM.nhdr \
-b 512 256 -t int -o $NUM-RGB-jh.nrrd
unu gamma -i $NUM-RGB-jh.nrrd -g 5 \
| unu heq -b 3000 -s 2 \
| unu flip -a 1 \
| unu quantize -b 8 -o $NUM-RGB-jh.png
end
end
After much experimentation, I determined the following steps which worked well to transform the 16-bit RGB values to 8-bit values. The steps are a little bizarre (very non-linear), but it seems to work on the available 48-bit slices. Basically, it increases saturation of colors away from the color of fat, and then applies a RGB-component-wise gamma. Boosting saturation was effective in increasing the color contrast between the different tissues, and between the synthetic yellow, green, and blue colors. However, this is not an operation that can be done with NRRD command-line tools, so I made a stand-alone executable to do this. Here is the source, as well as statically-linked pre-compiled versions for darwin, linux, and irix6 (64-bit).
foreach N ( 24 25 26 27 28 29 30 31 32 33 34 35 )
echo ${N}00-hl.png
gzcat ${N}00.48.raw.gz | ./makeHLI - ${N}00-hl.png
unu resample -i ${N}00-hl.png -s = x0.4 x0.4 -k cubic:0,0.5 \
-o ${N}00-hl-small.png &
end
unu join -i ??00-hl-small.png -a 3 \
| unu resample -k cubic:0,0.5 -s = 290 210 = \
| unu pad -min 0 -2 -2 0 -max 2 M+2 M+2 M -b pad -v 0 \
| unu reshape -s 3 294 214 3 4 \
| unu permute -p 0 1 3 2 4 \
| unu reshape -s 3 882 856 -o hl-test.png