|
|
|
|
|
International Workshop on Topological Data Analysis in Biomedicine (TDA-Bio)
Part of the 7th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM-BCB) |
|
|
|
Overview |
|
|
|
Biomedical Informatics in the Big Data Era Recently, a suite of new techniques termed topological data analysis (TDA) has shown a lot of promise in discovering structure in large, high-dimensional, and diverse data sets that other traditional techniques could not find. The range of applications includes gene expression analysis, voting, and basketball players' performances, to name a few. This workshop will present a concise yet self-contained overview of the key aspects of TDA, with an eye toward motivating the application of these techniques to problems in bioinformatics and computational biology (BCB). While topological techniques have been applied previously in certain subfields of BCB (e.g., to model protein and DNA/RNA 3D structure), they have proved to be much more versatile and powerful than these applications might suggest. We aim to showcase the versatility and strength of this suite of techniques in this workshop.
Why Topology? There are many important properties of topology that make efficient extraction of patterns from large data sets possible. First, topology studies shapes in a coordinate free way. In other words, topological constructions will not depend on the coordinate system chosen, but only on the distances between points in the data set. This will enable comparison among data sets derived from different platforms or coordinate systems. Second, topological constructions are not sensitive to small changes in data, and are robust against noise. Third, topology works with compressed representations of spaces in the form of simplicial complexes (e.g., triangulations), which can be viewed as a form of compression that preserves information relevant to how points are connected. Topological methods are also known to be more sensitive to both large and small scale patterns than other more traditional techniques such as principal component analysis (PCA), multidimensional scaling (MDS), and cluster analysis. Further, the "shapes" of the topological representations (simplicial complexes in general) naturally lend themselves to insightful visualization.
The Workshop |
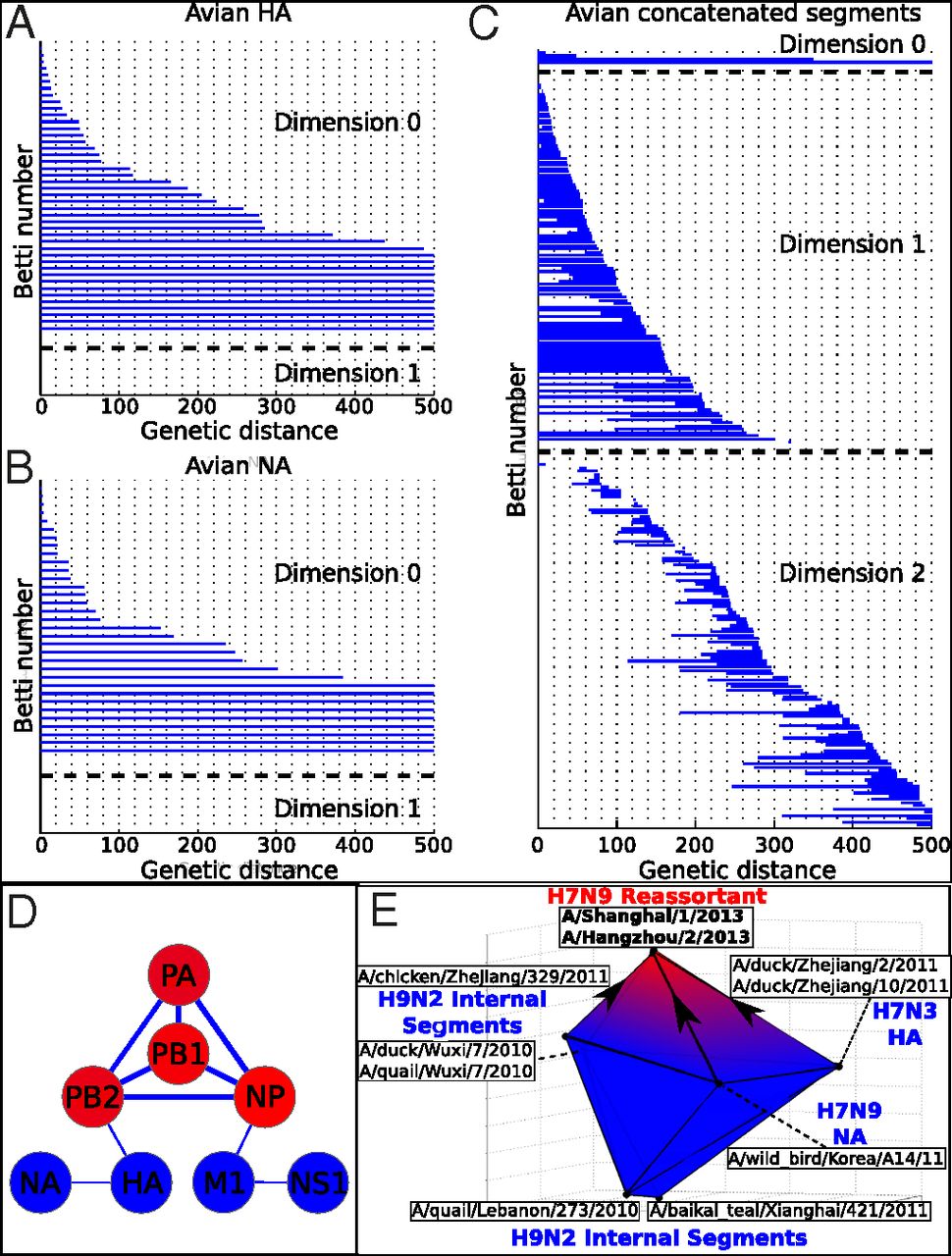 (source: PNAS) 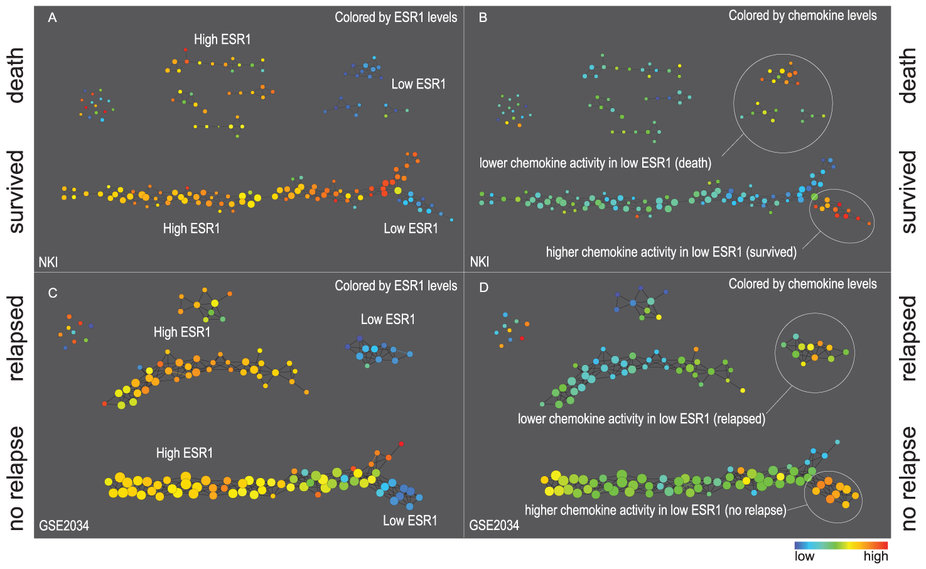 (source: Nature Scientific Reports) |
|
|
|
Tentative Schedule |
|
|
|
Since TDA is a relatively new area to the ACM-BCB audience, our plan will be to maximize the involvement of the audience in the workshop. To this end, we plan a full day format, with two sessions. With an eye toward increasing the exposure to students and junior researchers, we plan to have a demo session. We will also have a panel discussion on the potential applications of TDA in the BCB domain. |
|
|
|
|
|
|---|---|---|---|
| 8:50 - 9:00 | Opening remarks | ||
| 9:00 - 9:50 | Keynote Talk 1 |
Yusu Wang Associate professor of computer science at the Ohio State University |
Two Examples of Application of Topological Methods in Neuron Data Analysis |
| 10:00 - 10:35 | Invited Talk 1 |
Chao Chen Assistant professor of computer science at City University of New York |
Extracting and Using Topological Structures in the Analysis of Biomedical Images |
| 10:35 - 10:50 | Break | ||
| 10:50 - 11:25 | Invited Talk 2 |
Elizabeth Munch Assistant professor of mathematics at University at Albany |
Utilizing Topological Data Analysis to Detect Periodicity |
| 11:30 - 12:05 | Invited Talk 3 |
Brittany Fasy Assistant professor of computer science at Montana State University |
Using Topological Data Analysis to Study Glandular Architecture |
| 12:05 - 13:30 | Lunch | ||
| 13:30 - 14:20 | Keynote Talk 2 |
Gunnar Carlsson Professor of mathematics at Stanford University President and co-founder of Ayasdi |
The Shape of Biomedical Data |
| 14:30 - 15:20 | Software Demo |
Svetlana Lockwood PostDoc Fellow, Washington State University |
Open Source Software for TDA |
| 15:20 - 15:25 | Break | ||
| 15:25 - 16:00 | Invited Talk 4 |
Bei Wang Phillips Assistant professor of computer science at the University of Utah |
Topological Data Analysis for Brain Networks |
| 16:00 - 16:35 | Invited Talk 5 |
Michael Robinson Assistant Professor of Applied Mathematics at the American University |
Finding Cross-Species Orthologs with Local Topology |
| 16:40 - 17:10 | Panel Discussion | ||
| 17:10 - 17:15 | Closing remarks |
|
|
|
Talk Abstract |
|
|
|
Keynote Talk 1Yusu WangTitle: Two Examples of Application of Topological Methods in Neuron Data Analysis Abstract: In this talk, I will describe two of our recent efforts in analyzing neuron structures via topological methods. The first topic is neuron shape comparison via persistent homology. Persistent homology is an important development in the field of applied and computational topology in the past 15 years. It provides a way to summarize an input domain the lens of a specific filtration of the domain. We show how the persistence summary can be used to compare neuron trees. The second topic is neuron reconstruction via Morse theory. We presend a framework to automatically extract neuron tree structures from 2D / 3D images with the help of discrete Morse theory. We will give some preliminary results in each of these two directions. This is joint work with Yanjie Li, Suyi Wang, Partha Mitra and Giorgio Ascoli. [Slides] [Talk Video] Keynote Talk 2Gunnar CarlssonTitle: The Shape of Biomedical Data Abstract: The life sciences produce data sets which are often complex, and are not easily addressed by standard algebraic methods of modeling. This situation calls for new methods of modeling, and one such is topological modeling, based on the mathematical subdiscipline of topology. Roughly speaking, topology studies shape and its higher dimensional analogues, and can be adapted to the setting of point clouds, where most data sets reside. In this talk, we will discuss this methodology with numerous examples. [Slides] [Talk Video] Invited Talk 1Chao ChenTitle: Extracting and Using Topological Structures in the Analysis of Biomedical Images Abstract: In this talk, we will demonstrate how topological structures can be extracted and used in the analysis of cardiac and neuron images. In these cases, existing segmentation methods are challenged by lack of shape priors and inhomogeneity of the appearance. We show how topological information can form novel global prior and be used in the segmentation model. In the second half, we show how topological structures can help the clustering of high-dimensional discrete data, e.g., DNA data. [Slides] [Talk Video] Invited Talk 2Elizabeth MunchTitle: Utilizing Topological Data Analysis to Detect Periodicity Abstract: The field of TDA has shown itself to be a very powerful tool for data anlysis, finding structure not easily detectible by other methods. In this talk, we will look at two applications of TDA to time series where it is necessary to quantify periodicity in the system. The ability for TDA to accept different types of input means that these data come as time series in a broad sense, mean that the output could be real numbers, images, higher dimensional values, etc. The first application comes from engineering, where chatter behavior in a turning process leads to the finished parts being unuseable. In this application, we use Takens embedding on the real-valued time series to obtain a point cloud which can be investigated using persistent homology. The second application comes from atmospheric science, where persistent homology applied to a time series of IR images of a hurricane gives quantification of a periodic behavior previously only qualitatively described by domain scientists. These applications show that the techniques presented can be used on domain from a wide range of domains, as well as having the potential to find more complex behavior than just periodicity. [Slides] [Talk Video] Invited Talk 3Brittany FasyTitle: Using Topological Data Analysis to Study Glandular Architecture Abstract: The current standard for prostate cancer grading is the Gleason score, a subjective rating system based on an analysis of high-level tissue architecture and glandular shape and organization. This analysis can be aided with tools from topological data analysis. In particular, we use persistence diagrams, intensity plots (or persistence images), landscapes, and silhouettes as descriptors of the biopsy slides. We will discuss preliminary results on comparing regions of pure Gleason grades 3, 4, and 5. Other biological applications of TDA we will briefly discuss are finding correlations between biofilms and quantifying the significance of bubbles in De Bruijn graphs. [Slides] Invited Talk 4Bei Wang PhillipsTitle: Topological Data Analysis for Brain Networks Abstract: In this talk, we present a novel method for analyzing the relationship between functional brain networks and behavioral phenotypes. Drawing from topological data analysis, we first extract topological features using persistent homology from functional brain networks that are derived from correlations in resting-state fMRI. Rather than fixing a discrete network topology by thresholding the connectivity matrix, these topological features capture the network organization across all continuous threshold values. We then propose to use a kernel partial least squares (kPLS) regression to statistically quantify the relationship between these topological features and behavior measures. The kPLS also provides an elegant way to combine multiple image features by using linear combinations of multiple kernels. In our experiments we test the ability of our proposed brain network analysis to predict autism severity from rs-fMRI. We show that combining correlations with topological features gives better prediction of autism severity than using correlations alone. [Slides] [Talk Video] Invited Talk 5Michael RobinsonTitle: Finding Cross-Species Orthologs with Local Topology Abstract: Functionally and genetically related proteins from different species are called "orthologs". Knowledge about well-studied proteins in one species can be transferred to their othologs in other species. Since proteins are best understood both in genetic and functional contexts -- both realized as networks -- the problem of finding pairs of orthologs is related to network alignment problems. Various methods for network alignment exist, but they are difficult to employ at scale and tend to prefer global structure at the expense of local structure in the network. This talk will present a novel multi-stage topological prefilter that reduces the search space for pairs of orthologs dramatically. We will focus our attention on networks of protein-protein interactions (PPI), which can be useful in predicting protein function or identifying possible causes of disease. Proteins within and across species can also be classified in common orthologous groups (COGs) based upon their inferred ancestry. Using these two networks and our prefilter, we discovered local homological and local spectral features of the flag complex on hybrid protein-protein and protein-gene networks that appears to detect certain classes of cross-species orthologs. [Slides] [Talk Video] Software DemoSvetlana LockwoodTitle: Open Source Software for TDA Abstract: Topological data analysis (TDA) is a new and vibrant research field. The application of TDA ranges over a variety of disciplines from biological and brain networks to image segmentation to phylogenetic trees. In this demo we present open source software for two most popular methods of topological data analysis. The first method is based on persistent homology and is used to study the shape and the connectivity of the data space. The second method follows from the Reeb graph construction and is commonly known as Mapper. We present case studies for both methods complete with examples and code. [Slides] [Talk Video] (Apologies for some technical issues associated the presentation.) [Demo Code] Panel Discussions[Video] |
|
|
|
Registration |
|
|
|
Please register through ACM-BCB. |
|
|
|
Graduate Student Travel Support |
|
|
|
We expect some funding from NSF (CCF-1654106) to support the participation
of graduate students in the workshop. We will be able to support the registration and travel of up to $1000 per person for eight student participates. |
|
|
|
Organizers |
|
|
|
Bala Krishnamoorthy |
|
Bei Wang Phillips |
|
|
|
Acknowledgment |
|
|
|
The graduate student travel grant is provided by the National Science Foundation CCF-1654106 . Any opinions, findings, and conclusions or recommendations expressed in this workshop are those of author(s)/speaker(s) and do not necessarily reflect the views of the National Science Foundation.
|
|
|