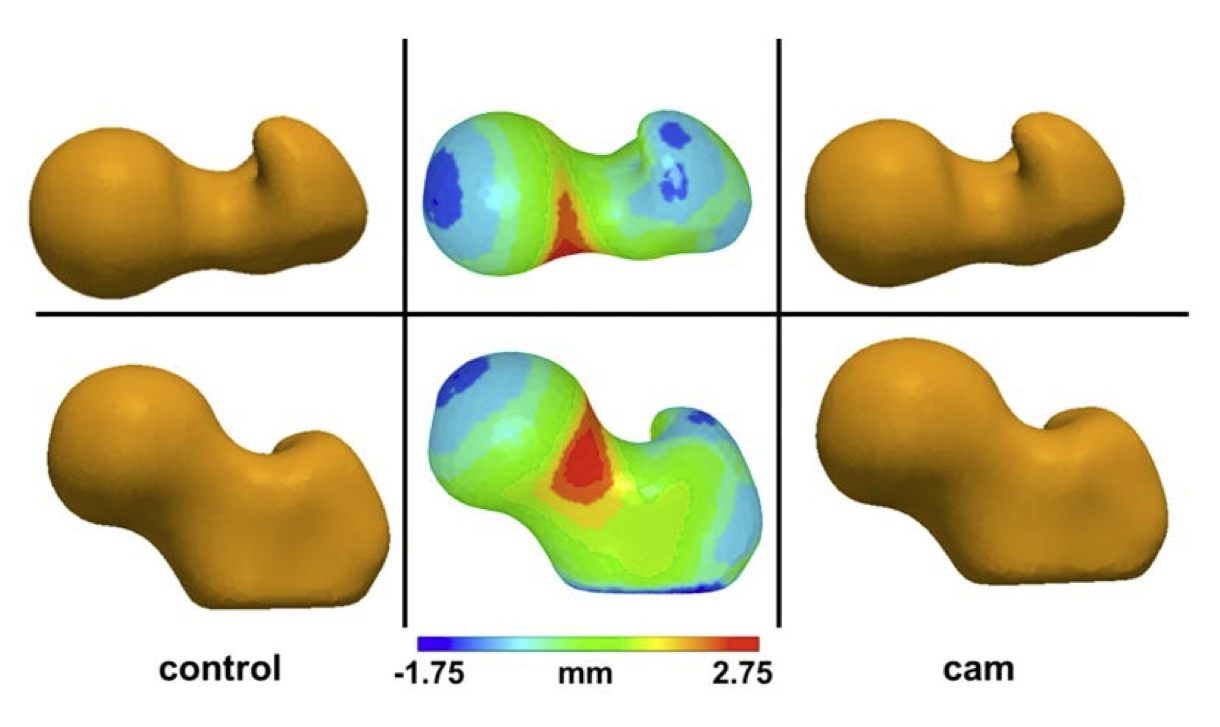
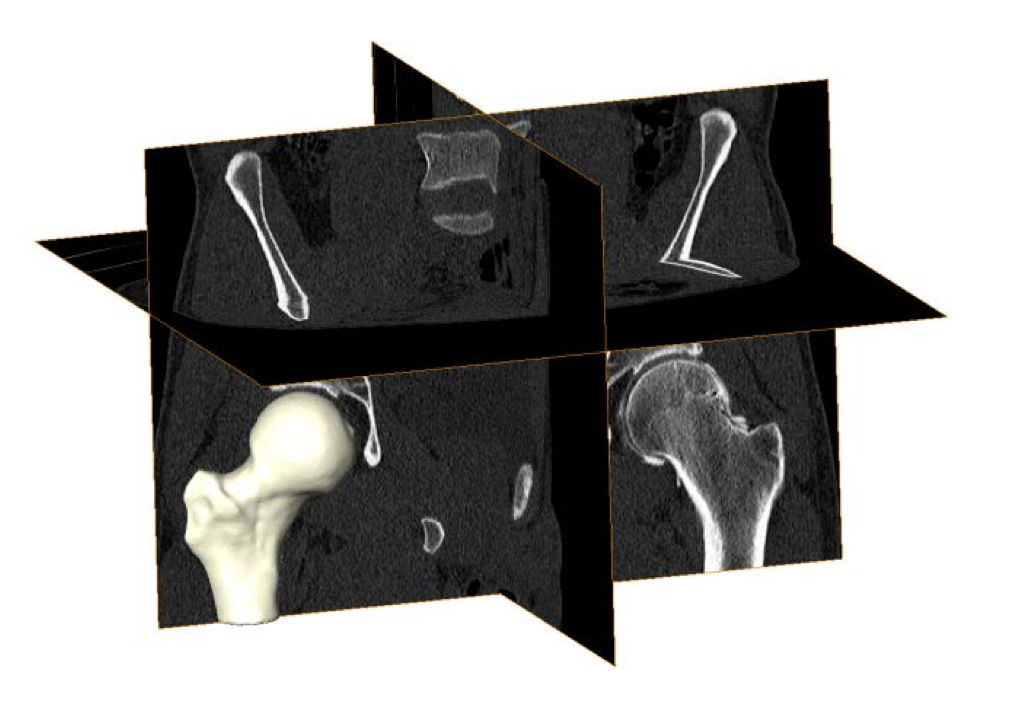
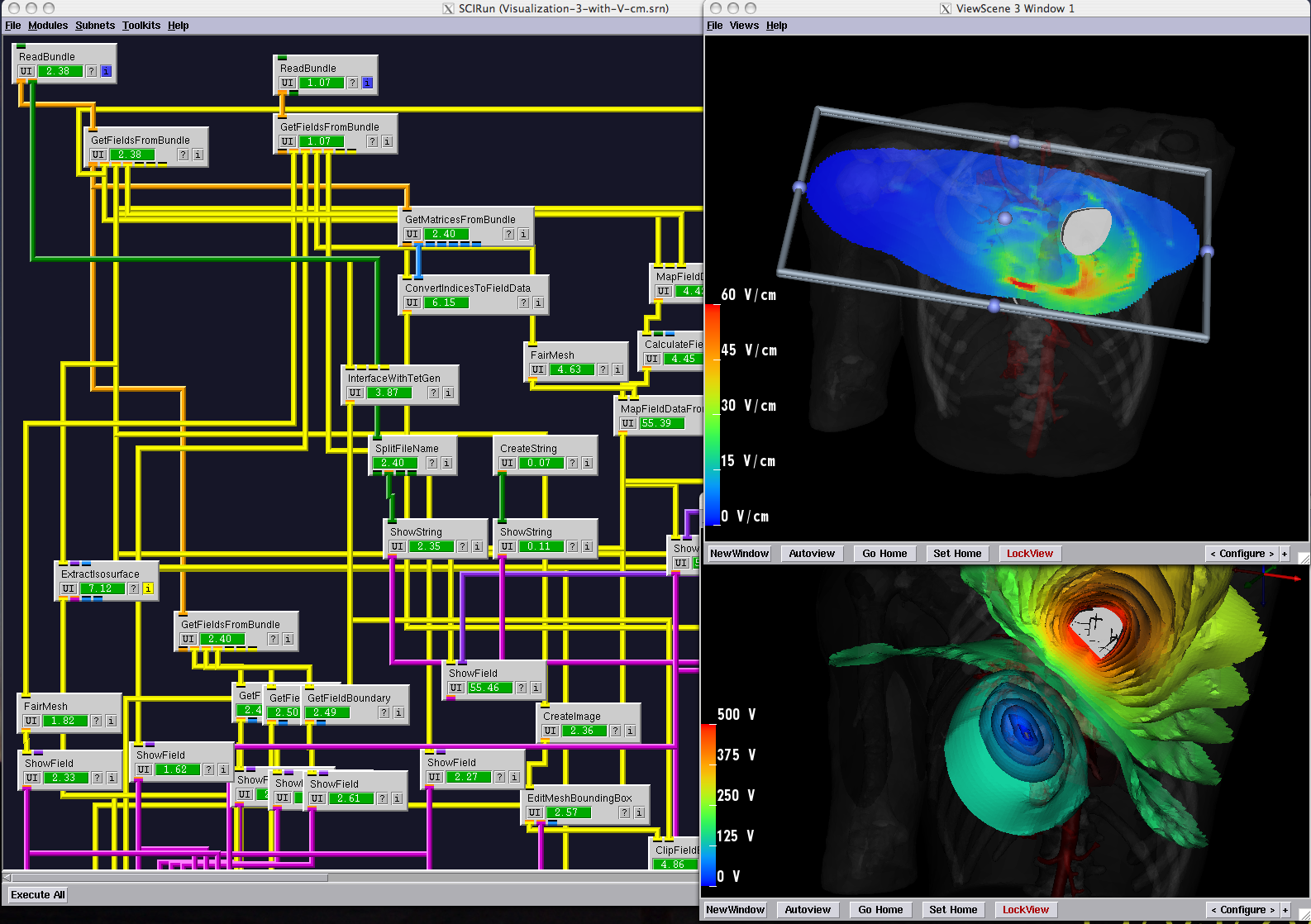
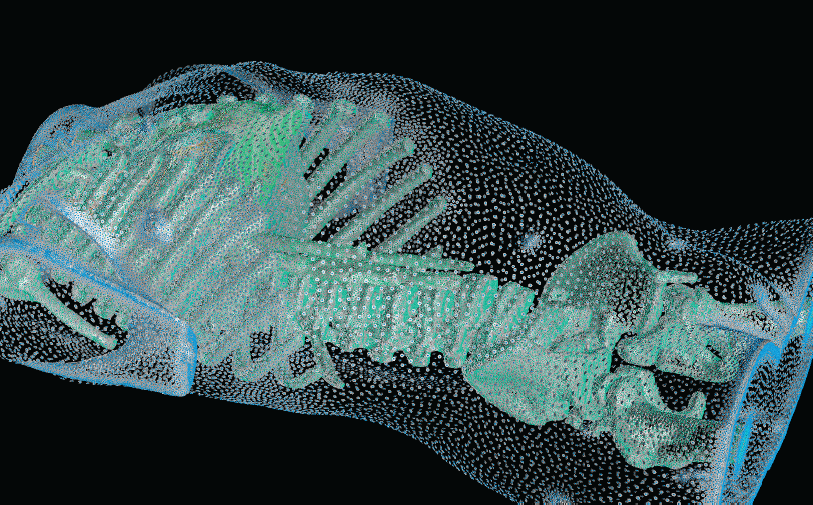
Cam Femoroacetabular Impingement Analysis using Statistical Shape Modeling
MRI Image Quantification Analysis for Atrial Fibrillation
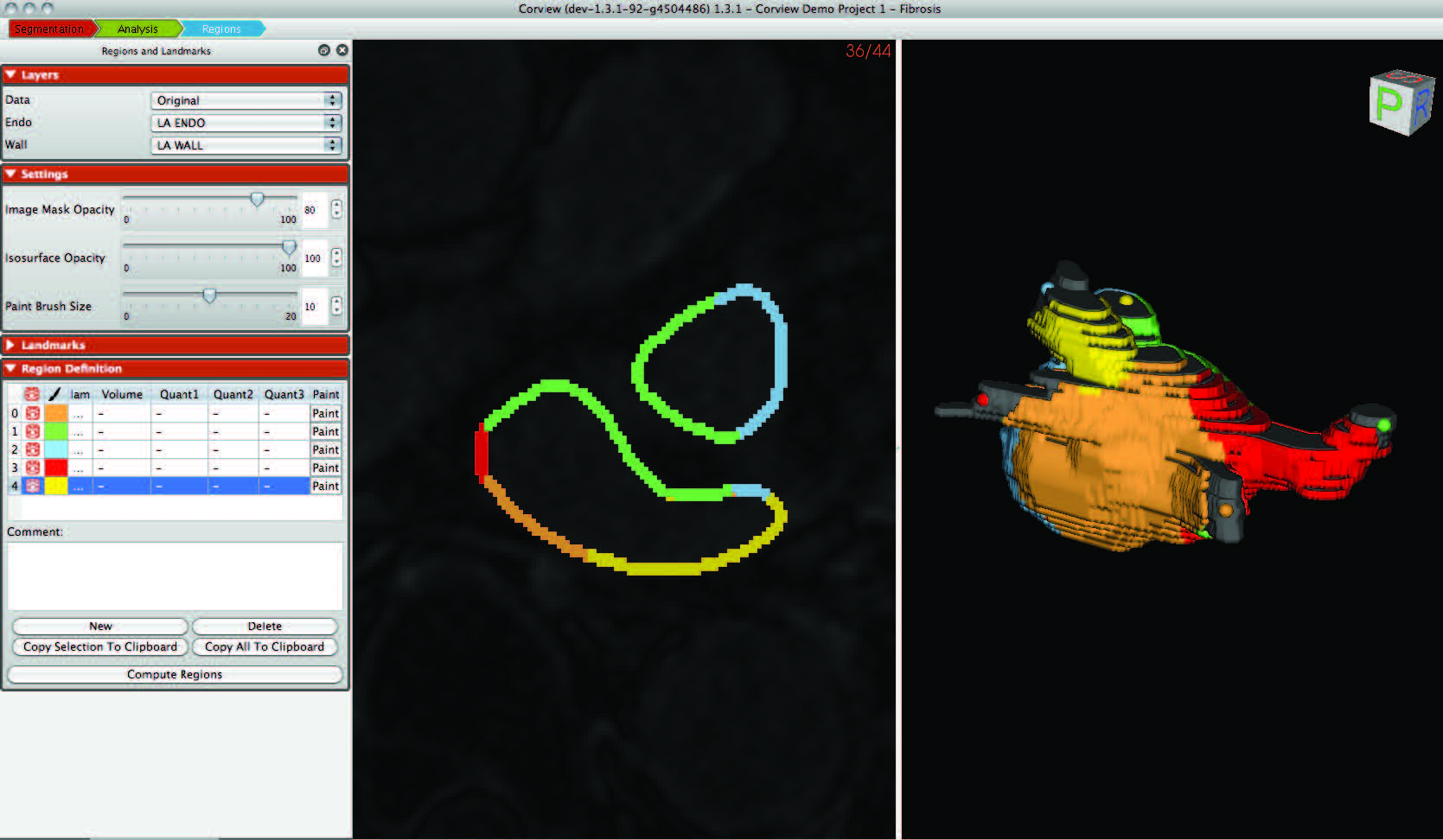 |
| Corview screenshot |
An interdisciplinary team at the Comprehensive Arrhythmia and MAnagement (CARMA) Center have made use of the segmentation, image analysis, and recently mesh generation and simulation capabilities of the CIBC to create a comprehensive program for AF management. The scope of the progress continues to expand each year and this application of CIBC technology has proven very fruitful even as it is very challenging.
Software Dissemination at the CIBC
Software Dissemination as a Form of Science and Technology Dissemination
The origins of our success in developing widely used software tools lie in a set of strategies for algorithm research and software development. One such strategy is the production of software tools with low barriers to entry. This entails the release of documented, tested, complete applications that do not require learning new programming languages or complex, architecture-specific build environments. We also continue to follow an initiative to create a suite of lightweight, stand-alone applications, directed at specific tasks of common interest across a wide set of disciplines. The result is a set of programs, such as Seg3D, with large and growing user bases.
Deep Brain Stimulation Planning with ImageVis3D
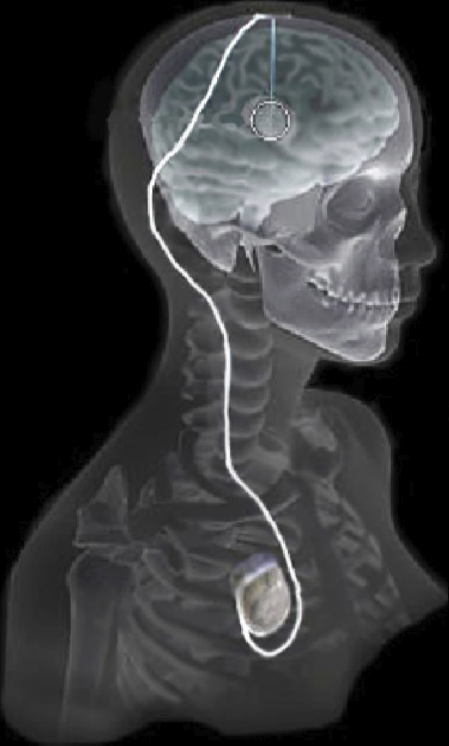 |
| Overview of the DBS system. The DBS electrode is implanted in the brain during stereotactic surgery. The electrode is attached via an extension wire to the IPG, which is implanted in the torso. The entire system is subcutaneous and is designed to deliver continuous stimulation for several years at a time. |
The selection of DBS settings is a significant clinical challenge that requires repeated revisions to achieve optimal therapeutic response, and is often performed without any visual representation of the stimulation system in the patient. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independent of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard care, but were selected in drastically less time. We found that our visualization system, available directly at the point of care on a device familiar to the clinician, can be used to guide clinical decision-making for selecting DBS settings. The positive impact of the system could also translate to areas other than DBS.
New Strategies in Biomedical Mesh Generation
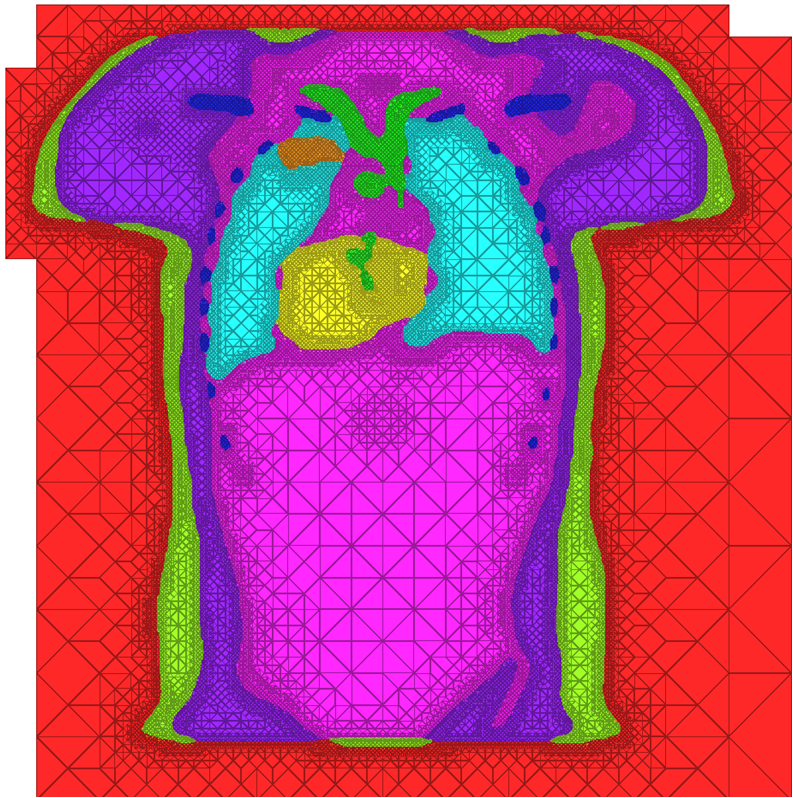 |
| A cross-section of a 3-dimensional, tetrahedral mesh of a torso. Each separate organ type is shown using a different color. |
High Quality Meshing
The problem of mesh generation has been widely studied, as a hybrid field of interest to the scientific, engineering, and computer science communities. In each of these fields, meshes are used to compute numerical approximations to solutions of partial differential equations. To do so, continuous mathematics are replaced with a discrete analogue, most commonly to facilitate the finite element method (FEM).The FEM works by decomposing a domain of interest into discrete entities of various dimensions, such as points (0-dimensional), edges (1-dimensional), and cells of higher dimension (frequently triangles and quadrilaterals are used for 2-dimensionl elements, tetrahedra and hexahedra for 3-dimensional). Together, these elements form what is commonly called a mesh (see figure)Solutions to the complex system are solved piecewise on each element, and then aggregated together to form the final solution. The FEM has become an important tool in medical imaging as well. For example, CT scans of legs can be meshed so that orthopedic modeling can accurately simulate gait, MRI scans of the torso are frequently used in cardiac electrophysical modeling, and images of the skull can identify structures of the brain.
Because the FEM is a computational tool that processes individual elements to approximate a whole solution, it is deeply impacted by the mesh elements used to represent the space. Two principle concerns stand out in the meshing problem for medical images:
Meshing for Multimaterial Biological Volumes: BioMesh3D
With the widespread use of medical imaging, there is a growing need for better analysis of datasets. One method for improving analysis is to simulate biological processes and medical interventions in silico, in order to render better predictions. For example, the CIBC center is currently collaborating with Dr. Triedman at Children's Hospital in Boston to develop a computer model that will help guide the implantation of Implantable Cardiac Defibrillators (ICDs). This model uses pediatric imaging to select placement of electrode leads to generate the optimal field for defibrillation. One of the critical pieces in the development of the model is the generation of quality meshes for electric field simulation. Because the project is entering the validation phase where many cases need to be reviewed, a robust and automated Meshing Pipeline is required.
Atrial Fibrillation
Uncertainty Visualization
 |
| An Isosurface visualization of a magnetic resonance imaging data set (in orange) surrounded by a volume rendered region of low opacity (in green) to indicate uncertainty in surface position. |
Imaging Meets Electrophysiology
In atrial fibrillation, the upper two chambers (the left and right atria) of the heart lose their synchronization and beat erratically and inefficiently. The same condition in the lower chambers (ventricles) of the heart is fatal within minutes and defibrillators are necessary to restore coordination. In the atria, death is by stealth and occurs over years, which is both good news and bad.
 Because it is not immediately fatal, there is time to treat atrial fibrillation–but also time to ignore it. While it is not immediately life-threatening, AF does immediately reduce the pumping capacity of the heart and elevates the heart rate of the entire organ. Patients cannot be as physically active as they often wish but many adjust to the symptoms and live with the disease untreated for many years.
Because it is not immediately fatal, there is time to treat atrial fibrillation–but also time to ignore it. While it is not immediately life-threatening, AF does immediately reduce the pumping capacity of the heart and elevates the heart rate of the entire organ. Patients cannot be as physically active as they often wish but many adjust to the symptoms and live with the disease untreated for many years.
Visualization
 |
| Figure from T. Fogal and J. Krüger, a Clearview rendering of the visible human male dataset |