SCI Publications
2012


C. Brownlee, J. Patchett, L.-T. Lo, D. DeMarle, C. Mitchell, J. Ahrens, C.D. Hansen.
“A Study of Ray Tracing Large-scale Scientific Data in Parallel Visualization Applications,” In Proceedings of the Eurographics Symposium on Parallel Graphics and Visualization (2012), Edited by H. Childs and T. Kuhlen and F. Marton, pp. 51--60. 2012.
Large-scale analysis and visualization is becoming increasingly important as supercomputers and their simulations produce larger and larger data. These large data sizes are pushing the limits of traditional rendering algorithms and tools thus motivating a study exploring these limits and their possible resolutions through alternative rendering algorithms . In order to better understand real-world performance with large data, this paper presents a detailed timing study on a large cluster with the widely used visualization tools ParaView and VisIt. The software ray tracer Manta was integrated into these programs in order to show that improved performance could be attained with software ray tracing on a distributed memory, GPU enabled, parallel visualization resource. Using the Texas Advanced Computing Center’s Longhorn cluster which has multi-core CPUs and GPUs with large-scale polygonal data, we find multi-core CPU ray tracing to be significantly faster than both software rasterization and hardware-accelerated rasterization in existing scientific visualization tools with large data.
Keywords: kaust, scidac


G. Chen, V. Kwatra, L.-Y. Wei, C.D. Hansen, E. Zhang.
“Design of 2D Time-Varying Vector Fields,” In IEEE Transactions on Visualization and Computer Graphics TVCG, Vol. 18, No. 10, pp. 1717--1730. 2012.
DOI: 10.1109/TVCG.2011.290


M. Kim, G. Chen, C.D. Hansen.
“Dynamic particle system for mesh extraction on the GPU,” In Proceedings of the 5th Annual Workshop on General Purpose Processing with Graphics Processing Units, London, England, GPGPU-5, ACM, New York, NY, USA pp. 38--46. 2012.
ISBN: 978-1-4503-1233-2
DOI: 10.1145/2159430.215943
Extracting isosurfaces represented as high quality meshes from three-dimensional scalar fields is needed for many important applications, particularly visualization and numerical simulations. One recent advance for extracting high quality meshes for isosurface computation is based on a dynamic particle system. Unfortunately, this state-of-the-art particle placement technique requires a significant amount of time to produce a satisfactory mesh. To address this issue, we study the parallelism property of the particle placement and make use of CUDA, a parallel programming technique on the GPU, to significantly improve the performance of particle placement. This paper describes the curvature dependent sampling method used to extract high quality meshes and describes its implementation using CUDA on the GPU.
Keywords: CUDA, GPGPU, particle systems, volumetric data mesh extraction


T. Martin, G. Chen, S. Musuvathy, E. Cohen, C.D. Hansen.
“Generalized Swept Mid-structure for Polygonal Models,” In Computer Graphics Forum, Vol. 31, No. 2 part 4, Wiley-Blackwell, pp. 805--814. May, 2012.
DOI: 10.1111/j.1467-8659.2012.03061.x
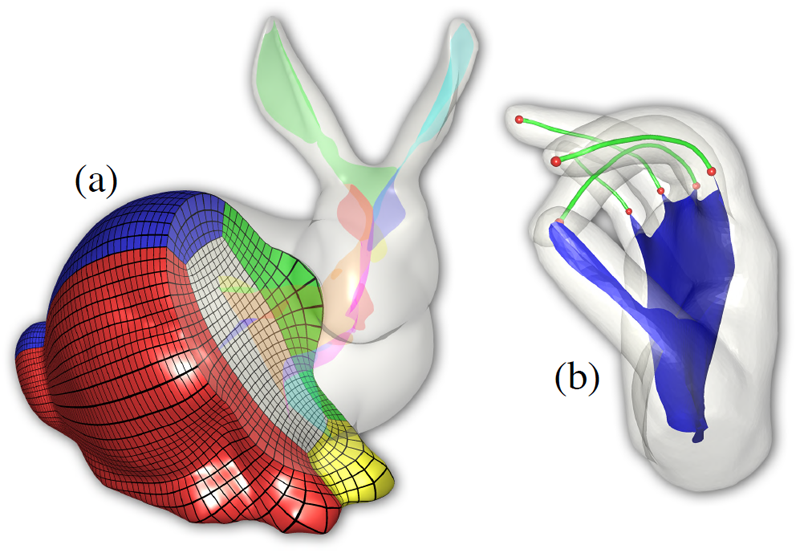
We introduce a novel mid-structure called the generalized swept mid-structure (GSM) of a closed polygonal shape, and a framework to compute it. The GSM contains both curve and surface elements and has consistent sheet-by-sheet topology, versus triangle-by-triangle topology produced by other mid-structure methods. To obtain this structure, a harmonic function, defined on the volume that is enclosed by the surface, is used to decompose the volume into a set of slices. A technique for computing the 1D mid-structures of these slices is introduced. The mid-structures of adjacent slices are then iteratively matched through a boundary similarity computation and triangulated to form the GSM. This structure respects the topology of the input surface model is a hybrid mid-structure representation. The construction and topology of the GSM allows for local and global simplification, used in further applications such as parameterization, volumetric mesh generation and medical applications.
Keywords: scidac, kaust


M. Schott, T. Martin, A.V.P. Grosset, C. Brownlee, Thomas Hollt, B.P. Brown, S.T. Smith, C.D. Hansen.
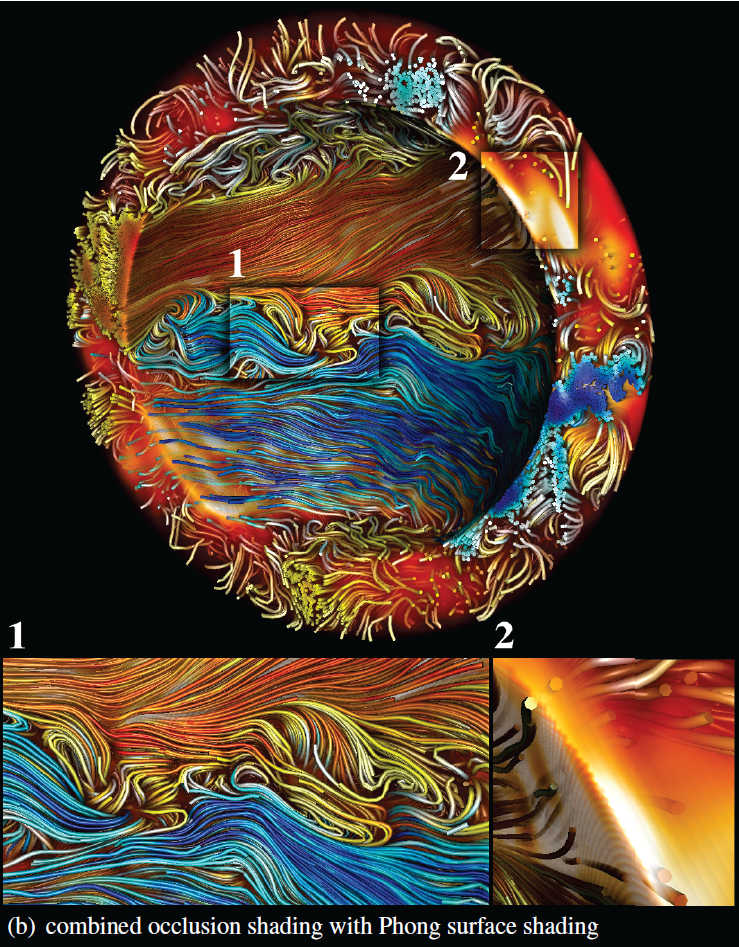
“Combined Surface and Volumetric Occlusion Shading,” In Proceedings of Pacific Vis 2012, pp. 169--176. 2012.
DOI: 10.1109/PacificVis.2012.6183588
In this paper, a method for interactive direct volume rendering is proposed that computes ambient occlusion effects for visualizations that combine both volumetric and geometric primitives, specifically tube shaped geometric objects representing streamlines, magnetic field lines or DTI fiber tracts. The proposed algorithm extends the recently proposed Directional Occlusion Shading model to allow the rendering of those geometric shapes in combination with a context providing 3D volume, considering mutual occlusion between structures represented by a volume or geometry.
Keywords: scidac, vacet, kaust, nvidia


Y. Wan, H. Otsuna, C.-B. Chien, C.D. Hansen.
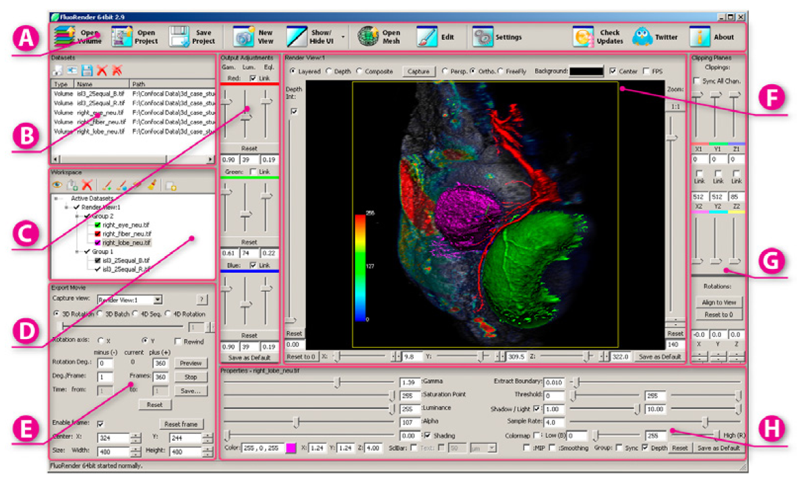
“FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research,” In Proceedings of Pacific Vis 2012, Incheon, Korea, pp. 201--208. 2012.
DOI: 10.1109/PacificVis.2012.6183592
2D image space methods are processing methods applied after the volumetric data are projected and rendered into the 2D image space, such as 2D filtering, tone mapping and compositing. In the application domain of volume visualization, most 2D image space methods can be carried out more efficiently than their 3D counterparts. Most importantly, 2D image space methods can be used to enhance volume visualization quality when applied together with volume rendering methods. In this paper, we present and discuss the applications of a series of 2D image space methods as enhancements to confocal microscopy visualizations, including 2D tone mapping, 2D compositing, and 2D color mapping. These methods are easily integrated with our existing confocal visualization tool, FluoRender, and the outcome is a full-featured visualization system that meets neurobiologists' demands for qualitative analysis of confocal microscopy data.
Keywords: scidac


Y. Wan, H. Otsuna, C.-B. Chien, C.D. Hansen.
“Interactive Extraction of Neural Structures with User-Guided Morphological Diffusion,” In Proceedings of the IEEE Symposium on Biological Data Visualization, pp. 1--8. 2012.
DOI: 10.1109/BioVis.2012.6378577
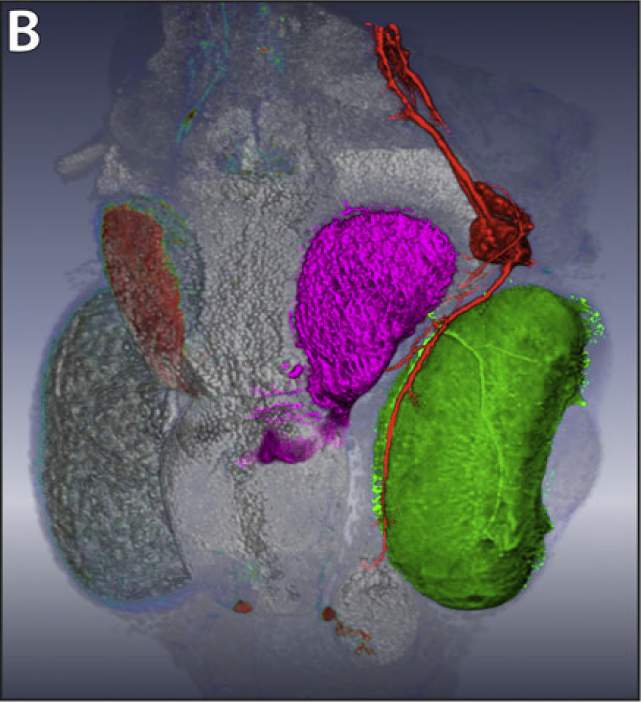
Extracting neural structures with their fine details from confocal volumes is essential to quantitative analysis in neurobiology research. Despite the abundance of various segmentation methods and tools, for complex neural structures, both manual and semi-automatic methods are ineffective either in full 3D or when user interactions are restricted to 2D slices. Novel interaction techniques and fast algorithms are demanded by neurobiologists to interactively and intuitively extract neural structures from confocal data. In this paper, we present such an algorithm-technique combination, which lets users interactively select desired structures from visualization results instead of 2D slices. By integrating the segmentation functions with a confocal visualization tool neurobiologists can easily extract complex neural structures within their typical visualization workflow.


Y. Wan, A.K. Lewis, M. Colasanto, M. van Langeveld, G. Kardon, C.D. Hansen.
“A Practical Workflow for Making Anatomical Atlases in Biological Research,” In IEEE Computer Graphics and Applications, Vol. 32, No. 5, pp. 70--80. 2012.
DOI: 10.1109/MCG.2012.64
An anatomical atlas provides a detailed map for medical and biological studies of anatomy. These atlases are important for understanding normal anatomy and the development and function of structures, and for determining the etiology of congenital abnormalities. Unfortunately, for biologists, generating such atlases is difficult, especially ones with the informative content and aesthetic quality that characterize human anatomy atlases. Building such atlases requires knowledge of the species being studied and experience with an art form that can faithfully record and present this knowledge, both of which require extensive training in considerably different fields. (For some background on anatomical atlases, see the related sidebar.)
With the latest innovations in data acquisition and computing techniques, atlas building has changed dramatically. We can now create atlases from 3D images of biological specimens, allowing for high-quality, faithful representations. Labeling of structures using fluorescently tagged antibodies, confocal 3D scanning of these labeled structures, volume rendering, segmentation, and surface reconstruction techniques all promise solutions to the problem of building atlases.
However, biology researchers still ask, \"Is there a set of tools we can use or a practical workflow we can follow so that we can easily build models from our biological data?\" To help answer this question, computer scientists have developed many algorithms, tools, and program codes. Unfortunately, most of these researchers have tackled only one aspect of the problem or provided solutions to special cases. So, the general question of how to build anatomical atlases remains unanswered.
For a satisfactory answer, biologists need a practical workflow they can easily adapt for different applications. In addition, reliable tools that can fit into the workflow must be readily available. Finally, examples using the workflow and tools to build anatomical atlases would demonstrate these resources' utility for biological research.
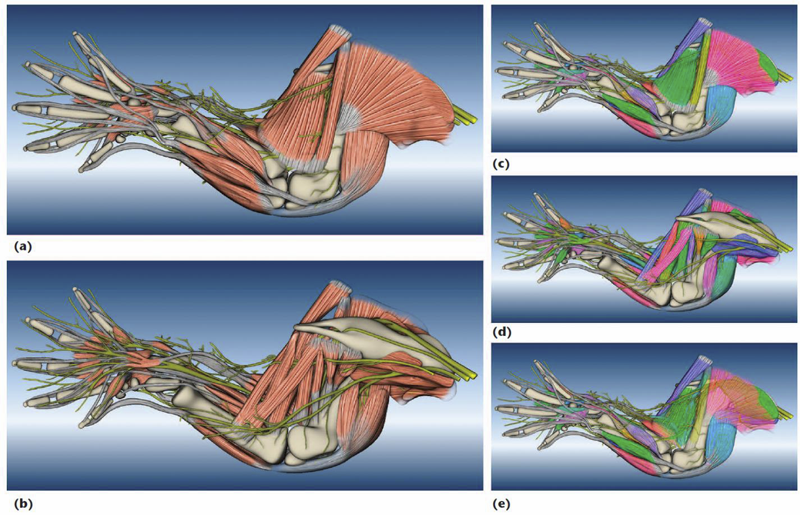
To build a mouse limb atlas for studying the development of the limb musculoskeletal system, University of Utah biologists, artists, and computer scientists have designed a generalized workflow for generating anatomical atlases. We adapted it from a CG artist's workflow of building 3D models for animated films and video games. The tools we used to build the atlas were mostly commercial, industry-standard software packages. Having been developed, tested, and employed for industrial use for decades, CG artists' workflow and tools, with certain adaptations, are the most suitable for making high-quality anatomical atlases, especially under strict budgetary and time limits. Biological researchers have been largely unaware of these resources. By describing our experiences in this project, we hope to show biologists how to use these resources to make anatomically accurate, high-quality, and useful anatomical atlases.


L. Zhou, M. Schott, C.D. Hansen.
“Transfer Function Combinations,” In Computers and Graphics, Vol. 36, No. 6, pp. 596--606. October, 2012.
DOI: 10.1016/j.cag.2012.02.007
Direct volume rendering has been an active area of research for over two decades. Transfer function design remains a difficult task since current methods, such as traditional 1D and 2D transfer functions are not always effective for all datasets. Various 1D or 2D transfer function spaces have been proposed to improve classification exploiting different aspects, such as using the gradient magnitude for boundary location and statistical, occlusion, or size metrics. In this paper, we present a novel transfer function method which can provide more specificity for data classification by combining different transfer function spaces. In this work, a 2D transfer function can be combined with 1D transfer functions which improve the classification. Specifically, we use the traditional 2D scalar/gradient magnitude, 2D statistical, and 2D occlusion spectrum transfer functions and combine these with occlusion and/or size-based transfer functions to provide better specificity. We demonstrate the usefulness of the new method by comparing to the following previous techniques: 2D gradient magnitude, 2D occlusion spectrum, 2D statistical transfer functions and 2D size based transfer functions.
Keywords: transfer function, volume rendering, classification, user interface, nih, scidac, kaust
2011


C. Brownlee, V. Pegoraro, S. Shankar, P.S. McCormick, C.D. Hansen.
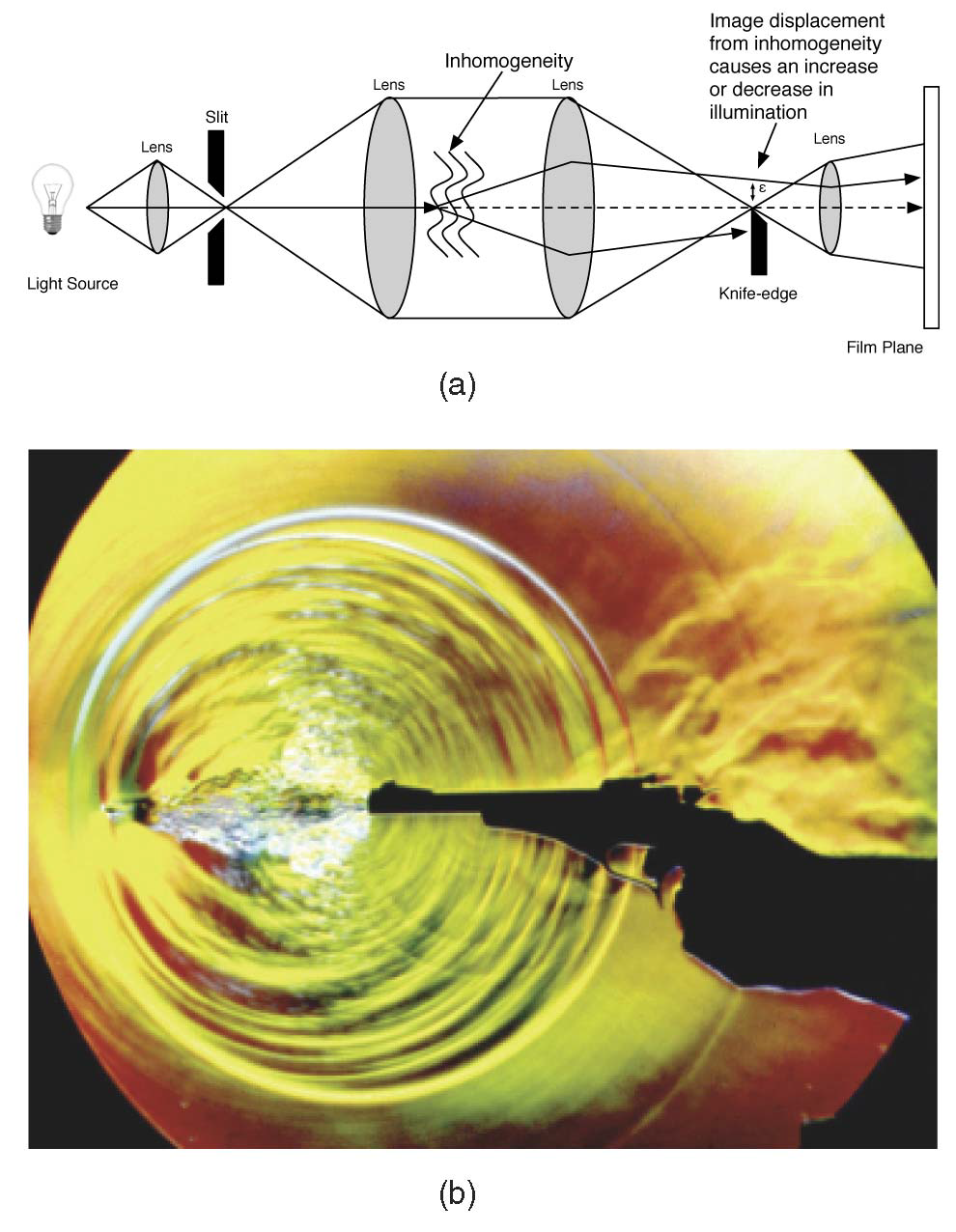
“Physically-Based Interactive Flow Visualization Based on Schlieren and Interferometry Experimental Techniques,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 17, No. 11, pp. 1574--1586. 2011.
Understanding fluid flow is a difficult problem and of increasing importance as computational fluid dynamics (CFD) produces an abundance of simulation data. Experimental flow analysis has employed techniques such as shadowgraph, interferometry, and schlieren imaging for centuries, which allow empirical observation of inhomogeneous flows. Shadowgraphs provide an intuitive way of looking at small changes in flow dynamics through caustic effects while schlieren cutoffs introduce an intensity gradation for observing large scale directional changes in the flow. Interferometry tracks changes in phase-shift resulting in bands appearing. The combination of these shading effects provides an informative global analysis of overall fluid flow. Computational solutions for these methods have proven too complex until recently due to the fundamental physical interaction of light refracting through the flow field. In this paper, we introduce a novel method to simulate the refraction of light to generate synthetic shadowgraph, schlieren and interferometry images of time-varying scalar fields derived from computational fluid dynamics data. Our method computes physically accurate schlieren and shadowgraph images at interactive rates by utilizing a combination of GPGPU programming, acceleration methods, and data-dependent probabilistic schlieren cutoffs. Applications of our method to multifield data and custom application-dependent color filter creation are explored. Results comparing this method to previous schlieren approximations are finally presented.
Keywords: uintah, c-safe


C. Brownlee, V. Pegoraro, S. Shankar, P.S. McCormick, C.D. Hansen.
“Physically-Based Interactive Flow Visualization Based on Schlieren and Interferometry Experimental Techniques,” In IEEE Transactions on Visualization and Computer Graphics, IEEE Transactions on Visualization and Computer Graphics, Vol. 17, No. 11, IEEE, pp. 1574--1586. November, 2011.
DOI: 10.1109/tvcg.2010.255


T. Ize, C.D. Hansen.
“RTSAH Traversal Order for Occlusion Rays,” In Computer Graphics Forum, Vol. 30, No. 2, Wiley-Blackwell, pp. 297--305. April, 2011.
DOI: 10.1111/j.1467-8659.2011.01861.x
We accelerate the finding of occluders in tree based acceleration structures, such as a packetized BVH and a single ray kd-tree, by deriving the ray termination surface area heuristic (RTSAH) cost model for traversing an occlusion ray through a tree and then using the RTSAH to determine which child node a ray should traverse first instead of the traditional choice of traversing the near node before the far node. We further extend RTSAH to handle materials that attenuate light instead of fully occluding it, so that we can avoid superfluous intersections with partially transparent objects. For scenes with high occlusion, we substantially lower the number of traversal steps and intersection tests and achieve up to 2x speedups.


T. Ize, C. Brownlee, C.D. Hansen.
“Real-Time Ray Tracer for Visualizing Massive Models on a Cluster,” In Proceedings of the 2011 Eurographics Symposium on Parallel Graphics and Visualization, pp. 61--69. 2011.
We present a state of the art read-only distributed shared memory (DSM) ray tracer capable of fully utilizing modern cluster hardware to render massive out-of-core polygonal models at real-time frame rates. Achieving this required adapting a state of the art packetized BVH acceleration structure for use with DSM and modifying the mesh and BVH data layouts to minimize communication costs. Furthermore, several design decisions and optimizations were made to take advantage of InfiniBand interconnects and multi-core machines.


A. Knoll, S. Thelen, I. Wald, C.D. Hansen, H. Hagen, M.E. Papka.
“Full-Resolution Interactive CPU Volume Rendering with Coherent BVH Traversal,” In Proceedings of IEEE Pacific Visualization 2011, pp. 3--10. 2011.


M. Schott, A.V.P. Grosset, T. Martin, V. Pegoraro, S.T. Smith, C.D. Hansen.
“Depth of Field Effects for Interactive Direct Volume Rendering,” In Computer Graphics Forum, Vol. 30, No. 3, Edited by H. Hauser and H. Pfister and J.J. van Wijk, Wiley-Blackwell, pp. 941--950. jun, 2011.
DOI: 10.1111/j.1467-8659.2011.01943.x
In this paper, a method for interactive direct volume rendering is proposed for computing depth of field effects, which previously were shown to aid observers in depth and size perception of synthetically generated images. The presented technique extends those benefits to volume rendering visualizations of 3D scalar fields from CT/MRI scanners or numerical simulations. It is based on incremental filtering and as such does not depend on any precomputation, thus allowing interactive explorations of volumetric data sets via on-the-fly editing of the shading model parameters or (multi-dimensional) transfer functions.
2010


C. Brownlee, V. Pegoraro, S. Shankar, P. McCormick, C.D. Hansen.
“Physically-Based Interactive Schlieren Flow Visualization,” In Proceedings of IEEE Pacific Visualization 2010, Note: Won Best Paper Award!, 2010.


Y. Hijazi, A. Knoll, M. Schott, A. Kensler, C.D. Hansen, H. Hagen.
“CSG Operations of Arbitary Primitives with Interval Arithmetic and Real-Time Ray Casting,” In DROPS - Dagstuhl Research Online Publication Server, In Scientific Visualization: Advanced Concepts, Edited by Hans Hagen, Schloss Dagstuhl - Leibniz-Zentrum fuer Informatik, pp. 78-89. 2010.


M. Schott, P. Grosset, T. Martin, V. Pegoraro, C.D. Hansen.
“Depth of Field Effects for Interactive Direct Volume Rendering,” SCI Technical Report, No. UUSCI-2010-005, SCI Institute, University of Utah, 2010.


Y. Wan, C.D. Hansen.
“Fast Volumetric Data Exploration with Importance-Based Accumulated Transparency Modulation,” In Proceedings of IEEE/EG International Symposium on Volume Graphics 2010, pp. 61--68. 2010.
DOI: 10.2312/VG/VG10/061-068
Direct volume rendering techniques have been successfully applied to visualizing volumetric datasets across many application domains. Due to the sensitivity of transfer functions and the complexity of fine-tuning transfer functions, direct volume rendering is still not widely used in practice. For fast volumetric data exploration, we propose Importance-Based Accumulated Transparency Modulation which does not rely on transfer function manipulation. This novel rendering algorithm is a generalization and extension of the Maximum Intensity Difference Accumulation technique. By only modifying the accumulated transparency, the resulted volume renderings are essentially high dynamic range. We show that by using several common importance measures, different features of the volumetric datasets can be highlighted. The results can be easily extended to a high-dimensional importance difference space, by mixing the results from an arbitrary number of importance measures with weighting factors, which all control the final output with a monotonic behavior. With Importance-Based Accumulated Transparency Modulation, the end-user can explore a wide variety of volumetric datasets quickly without the burden of manually setting and adjusting a transfer function.
2009


E.W. Bethel, C.R. Johnson, S. Ahern, J. Bell, P.-T. Bremer, H. Childs, E. Cormier-Michel, M. Day, E. Deines, P.T. Fogal, C. Garth, C.G.R. Geddes, H. Hagen, B. Hamann, C.D. Hansen, J. Jacobsen, K.I. Joy, J. Krüger, J. Meredith, P. Messmer, G. Ostrouchov, V. Pascucci, K. Potter, Prabhat, D. Pugmire, O. Rubel, A.R. Sanderson, C.T. Silva, D. Ushizima, G.H. Weber, B. Whitlock, K. Wu.
“Occam's Razor and Petascale Visual Data Analysis,” In Journal of Physics: Conference Series, Journal of Physics: Conference Series, Vol. 180, No. 012084, pp. (published online). 2009.
DOI: 10.1088/1742-6596/180/1/012084
One of the central challenges facing visualization research is how to effectively enable knowledge discovery. An effective approach will likely combine application architectures that are capable of running on today's largest platforms to address the challenges posed by large data with visual data analysis techniques that help find, represent, and effectively convey scientifically interesting features and phenomena.