Technical Collaboration for Brain Source Localization Software
Carsten Wolters, Ph.D.
Institute for Biomagnetism and Biosignal Analysis, University of Münster
Background
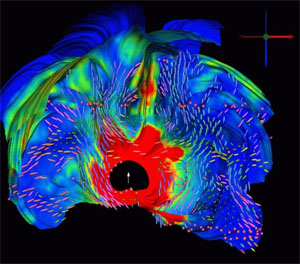 |
| Current flow from similated potential source in brain volume |
This collaboration represents a long standing relationship between Dr. Wolters and his group and the Center that has resulted in many publications together as well as shared progress in technological and software development. Dr. Wolters is a member of a very productive brain research group in Germany and has access to state of the art behavioral measurement systems for EEG and MEG, which he and his group use to perform many studies of event-related fields and potentials, functional organization and reorganization of cortical structures, the neurophysiological foundation of music, and MEG-fMRI-integration on human subjects. Dr. Wolters has also coordinated and participated in the development of a large scale software system, SimBio. SimBio is a generic environment for bio-numerical simulations, and is a freely available open-source software toolbox that focuses on carrying out source analysis in the human brain from electroencephalography (EEG), magnetoencephalography (MEG and magnetic resonance imaging (MRI) data. SimBio is a pure computational code, one part of it was already integrated as a module in the SCIRun software. Our goal, SCIRun-SimBio, is a code that combines the features of both software packages. MRI registration and segmentation and FE meshing is done within SCIRun, SimBio is then called for source analysis and the results of this pipeline are visualized within SCIRun.
Aims
The spectrum of collaborative aims with Dr. Wolters is very broad and includes both technical and neurological aspects of his research. We propose to focus special attention on the following aims:
- Interfacing of SCIRun and SimBio so that users of SCIRun can have access to the outstanding computational support for brain source problems that is contained in SimBio. In this setting, SCIRun would contains the tools required to create image based models and to visualize results of the simulations.
- The evaluation of diverse strategies for the construction of patient specific BEM and FEM geometric models of the head for use in bioelectric source localization problems in the brain. SimBio also has support for mesh generation and Dr. Wolters has head models that have been optimized over many years, against which we can compare the results of BioMesh3D.
- Implementation of brain source localization schemes using streaming architectures. The GPU based computing that will emerge from the Center will also be of great interest to brain source localization as the most common approaches in this field all involve repetitive solving of forward simulations over potentially large geometric meshes of both regular (hexahedral) and irregular (tetrahedral) structure. Acceleration of these tasks by means of GPU based computing would have a dramatic effect on the clinical utility of the approach.
- Applying polynomial chaos based parameter sensitivity analysis to brain source problems. The approaches for carrying out statistical parameter sensitivity we will implement in SCIRun have been applied to applications of cardiac bioelectricity but not yet in the brain. The collaboration will facilitate the evaluation of polynomial chaos approaches in this new domain and provide novel means of assessing the accuracy and robustness of source localization approaches in a wide range of clinical applications provided by Dr. Wolters.
Publications
- S. Lew, C.H. Wolters, A. Anwander, S. Makeig, and R.S. Macleod. "Improved eeg source analysis using low-resolution conductivity estimation in a four-compartment finite element head model." Hum Brain Mapp, 30(9):2862–2878, Dec 2009. http://dx.doi.org/10.1002/hbm.20714.
- S. Lew, C.H. Wolters, T. Dierkes, C. Roer, and R.S. MacLeod. "Accuracy and run-time comparison for different potential approaches and iterative solvers in finite element method based eeg source analysis." Applied Num. Math., 59:1970–1988, 2009.
- C.H. Wolters, A. Anwander, X. Tricoche, D.M. Weinstein, M.A. Koch, and R.S. Macleod. "Influence of tissue conductivity anisotropy on eeg/meg field and return current computation in a realistic head model: A simulation and visualization study using high-resolution finite element modeling." Neuroimage, 30(3):813–826, 2006.
- C.H. Wolters, A. Anwander, X. Tricoche, D. Weinstein, M.A. Koch, R.S. MacLeod. "Influence of Tissue Conductivity Anisotropy on EEG/MEG Field and Return Current Computation in a Realistic Head Model: A Simulation and Visualization Study Using High-Resolution Finite Element Modeling" In Neuroimage, December, 2005.
- C. H. Wolters, A. Anwander, B. Maess, R. S. MacLeod, and A. D. Friederici. "The influence of volume conduction effects on the EEG/MEG reconstruction of the sources of the Early Left Anterior Negativity." In Proc. of the 26th Annual Int. Conf. IEEE Engineering in Medicine and Biology Society, pages 3569– 3572, San Francisco, USA, Sep. 1-5, http://www.ucsfresno.edu/embs2004, 2004.

