SCIRun 5 Development
There must be considerable motivation for such a major release, motivation which comes from both our users, collaborators, and DBP partners but also from advances in software engineering and scientific computing, with which we must also keep pace. Our users continue to demand more efficiency, more flexibility in programming the workflows created with SCIRun, more support for big data, and more transparent access to large compute resources when simulations exceed the useful capacity of local resources. The evolution of software engineering has led to changes in computer languages, programming paradigms, visualization hardware and processing, user interface design (and tools to support this critical component), and the third party libraries that form the building blocks of complex scientific software. SCIRun 5 is a response to all these changing conditions and needs and also represents some long awaited refactoring that will provide greater flexibility and freedom as we move into the next generation of scientific computing.
Mesh Generation and Cleaver
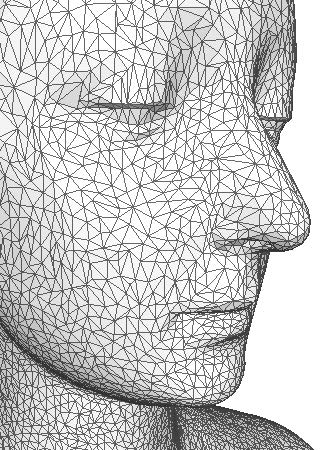 Figure 1: 3D surface mesh of a face. |
Evolution of the Medical Classroom
 Introducing ViSOAR. As data acquisition advances, and data sizes increase, the need for tools to process and visualize the results in an effective and efficient manner is becoming increasingly important. The reliance on supercomputers for scientific visualization and analysis is already proving to be a hindrance for wide accessibility to researchers and scientists dealing with large data.
Introducing ViSOAR. As data acquisition advances, and data sizes increase, the need for tools to process and visualize the results in an effective and efficient manner is becoming increasingly important. The reliance on supercomputers for scientific visualization and analysis is already proving to be a hindrance for wide accessibility to researchers and scientists dealing with large data.The Scientific Computing and Imaging (SCI) Institute and the Center for Extreme Data Management, Analysis, and Visualization (CEDMAV), in collaboration with ARUP Laboratories and the University of Utah, Department of Neurobiology and Anatomy, have developed ViSOAR--a multi platform visualization application for accessing and processing very large imaging data.
Neural Bioelectricity with EGI
 In collaboration with Dr. Don Tucker and his colleagues at Electrical Geodesics Inc (EGI) and the University of Oregon, this DBP is concerned with improving our ability to reconstruct and visualize neuroelectric sources (source localization) from EEG measurements and also our ability to stimulate specific brain regions using electrodes attached on to the scalp of the subject (transcranial direct current stimulation, tDCS).
In collaboration with Dr. Don Tucker and his colleagues at Electrical Geodesics Inc (EGI) and the University of Oregon, this DBP is concerned with improving our ability to reconstruct and visualize neuroelectric sources (source localization) from EEG measurements and also our ability to stimulate specific brain regions using electrodes attached on to the scalp of the subject (transcranial direct current stimulation, tDCS).For both research and clinical practice, EEG is a cost-effective tool to understand and excite brain activity. EEG advances have significantly improved the spatial resolution of source estimates and offer the promise of precise spatio-temporal monitoring and stimulation of cortical brain activity. By itself, high-resolution EEG would be affordable even for small hospitals in remote locations and could be easily managed by technicians in the field.
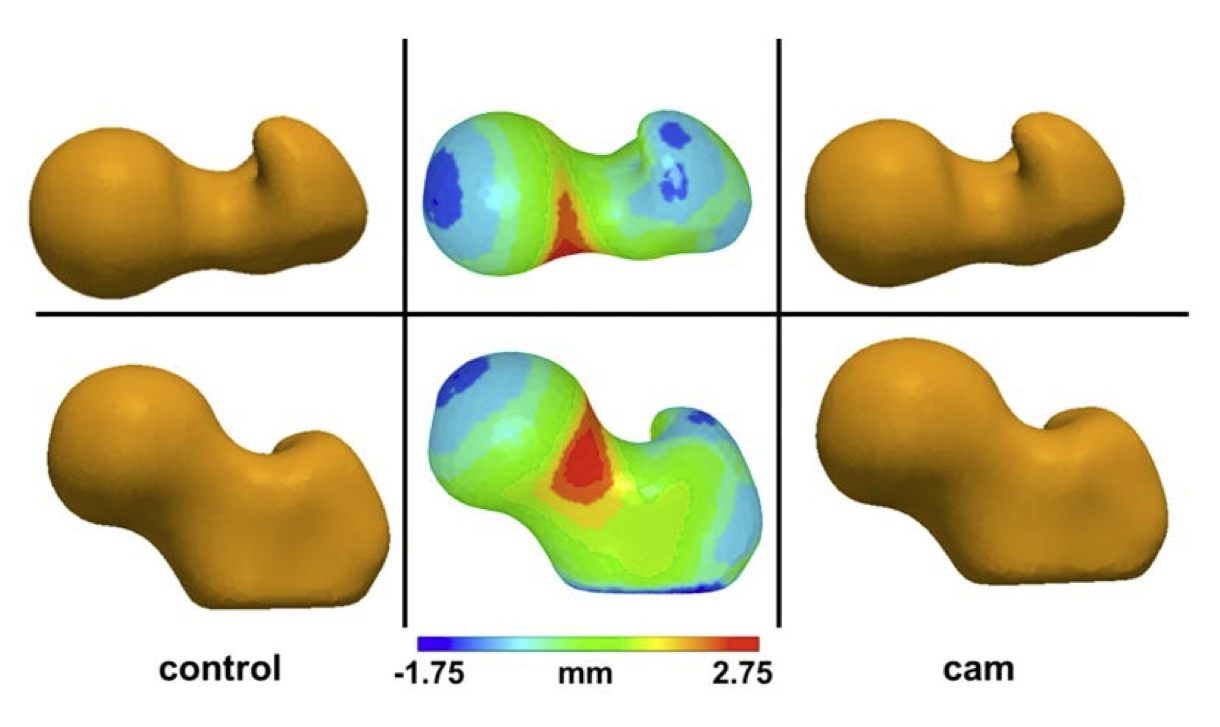
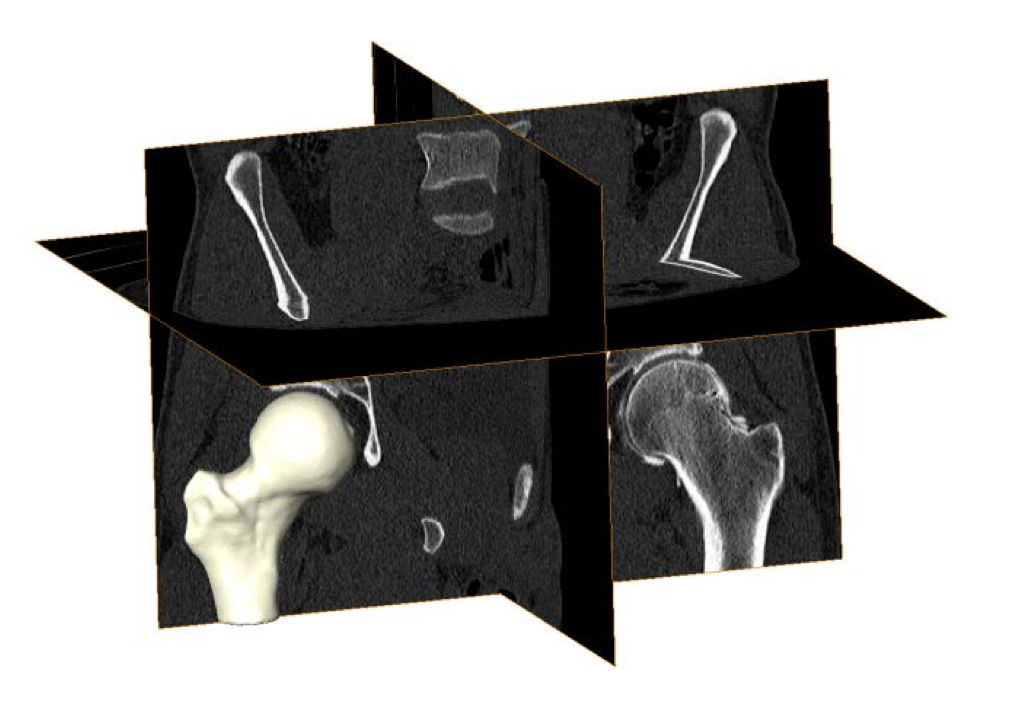
Cam Femoroacetabular Impingement Analysis using Statistical Shape Modeling
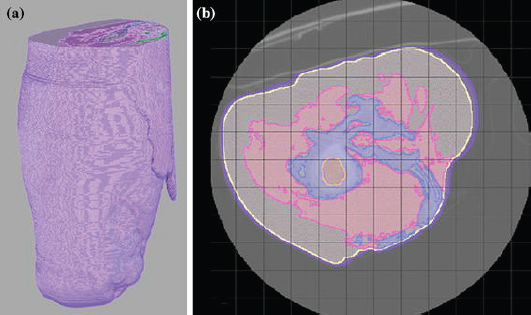
MRI Image Quantification Analysis for Atrial Fibrillation
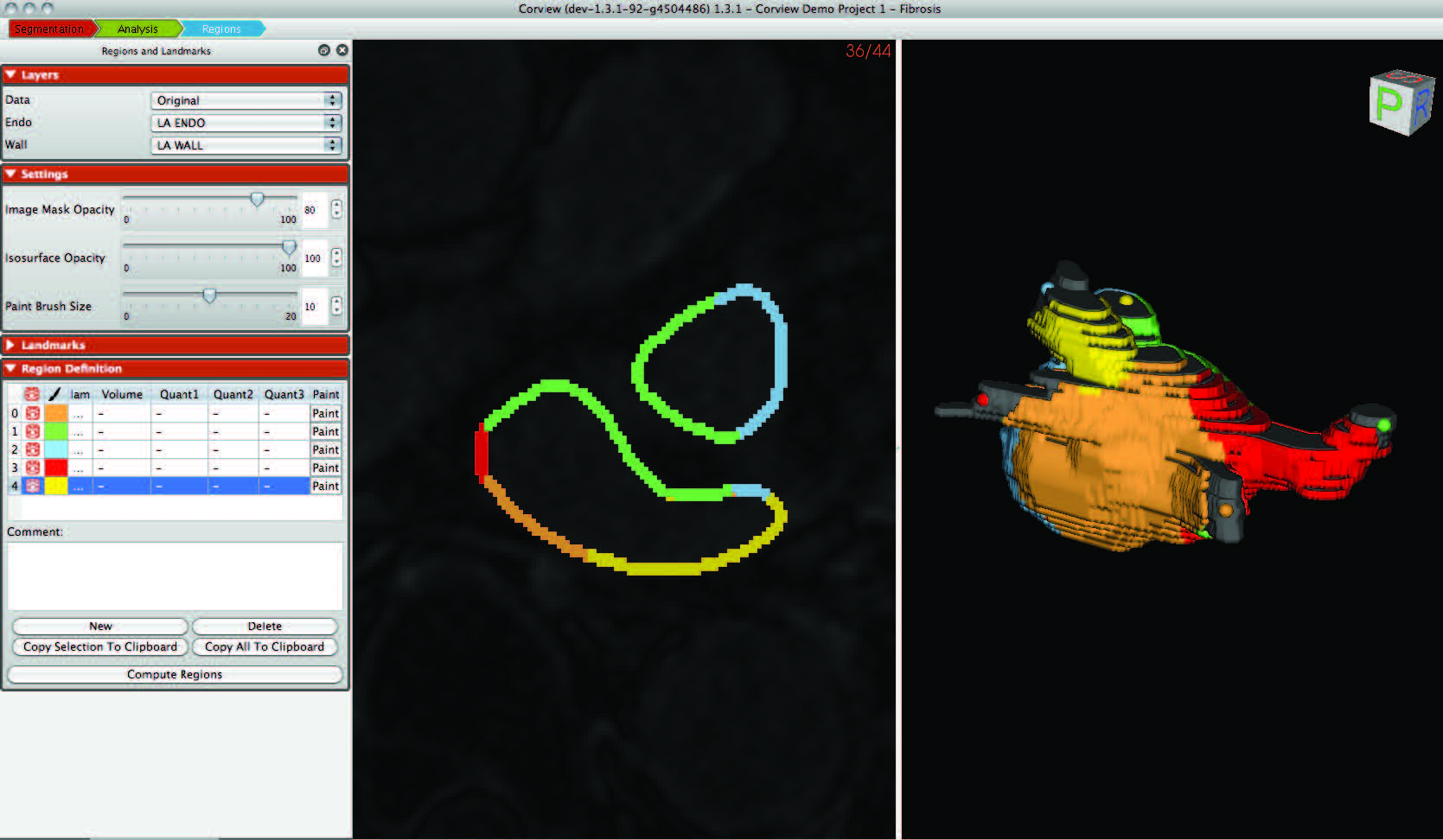 |
| Corview screenshot |
An interdisciplinary team at the Comprehensive Arrhythmia and MAnagement (CARMA) Center have made use of the segmentation, image analysis, and recently mesh generation and simulation capabilities of the CIBC to create a comprehensive program for AF management. The scope of the progress continues to expand each year and this application of CIBC technology has proven very fruitful even as it is very challenging.
Deep Brain Stimulation Planning with ImageVis3D
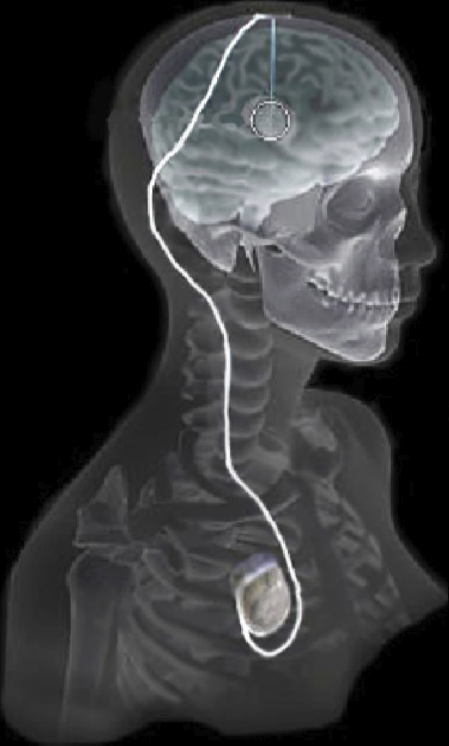 |
| Overview of the DBS system. The DBS electrode is implanted in the brain during stereotactic surgery. The electrode is attached via an extension wire to the IPG, which is implanted in the torso. The entire system is subcutaneous and is designed to deliver continuous stimulation for several years at a time. |
The selection of DBS settings is a significant clinical challenge that requires repeated revisions to achieve optimal therapeutic response, and is often performed without any visual representation of the stimulation system in the patient. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independent of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard care, but were selected in drastically less time. We found that our visualization system, available directly at the point of care on a device familiar to the clinician, can be used to guide clinical decision-making for selecting DBS settings. The positive impact of the system could also translate to areas other than DBS.
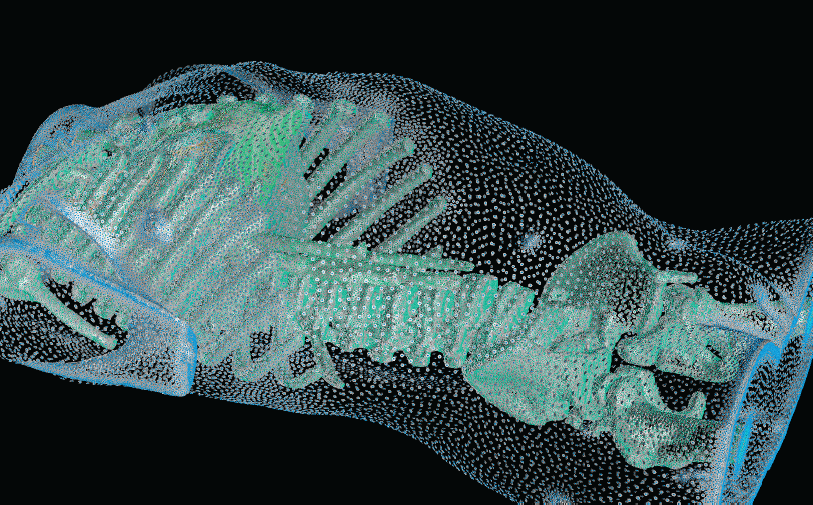
New Strategies in Biomedical Mesh Generation
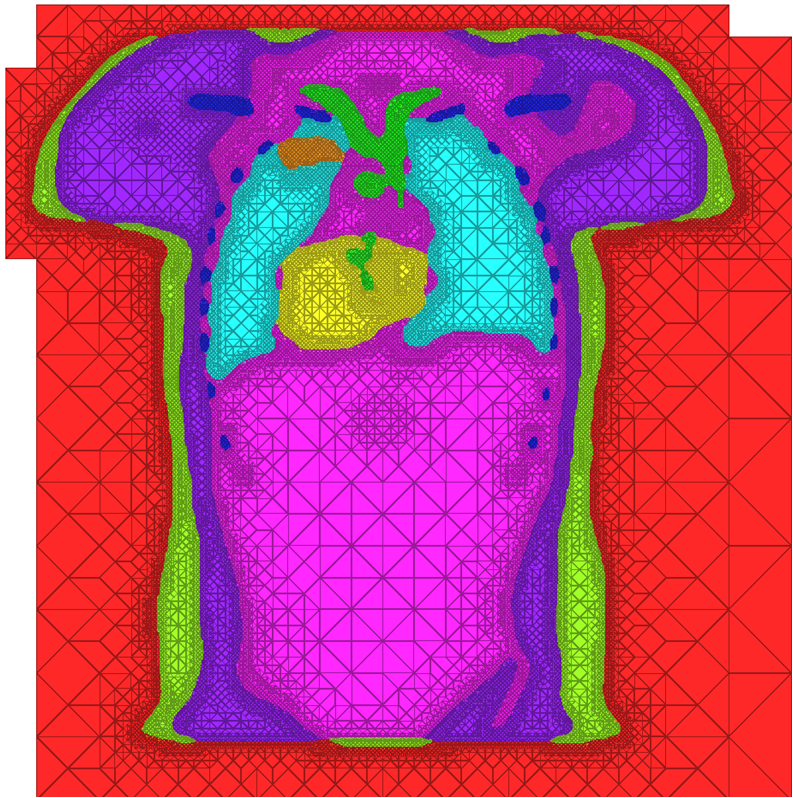 |
| A cross-section of a 3-dimensional, tetrahedral mesh of a torso. Each separate organ type is shown using a different color. |
High Quality Meshing
The problem of mesh generation has been widely studied, as a hybrid field of interest to the scientific, engineering, and computer science communities. In each of these fields, meshes are used to compute numerical approximations to solutions of partial differential equations. To do so, continuous mathematics are replaced with a discrete analogue, most commonly to facilitate the finite element method (FEM).The FEM works by decomposing a domain of interest into discrete entities of various dimensions, such as points (0-dimensional), edges (1-dimensional), and cells of higher dimension (frequently triangles and quadrilaterals are used for 2-dimensionl elements, tetrahedra and hexahedra for 3-dimensional). Together, these elements form what is commonly called a mesh (see figure)Solutions to the complex system are solved piecewise on each element, and then aggregated together to form the final solution. The FEM has become an important tool in medical imaging as well. For example, CT scans of legs can be meshed so that orthopedic modeling can accurately simulate gait, MRI scans of the torso are frequently used in cardiac electrophysical modeling, and images of the skull can identify structures of the brain.
Because the FEM is a computational tool that processes individual elements to approximate a whole solution, it is deeply impacted by the mesh elements used to represent the space. Two principle concerns stand out in the meshing problem for medical images:
Software Dissemination at the CIBC
Software Dissemination as a Form of Science and Technology Dissemination
The origins of our success in developing widely used software tools lie in a set of strategies for algorithm research and software development. One such strategy is the production of software tools with low barriers to entry. This entails the release of documented, tested, complete applications that do not require learning new programming languages or complex, architecture-specific build environments. We also continue to follow an initiative to create a suite of lightweight, stand-alone applications, directed at specific tasks of common interest across a wide set of disciplines. The result is a set of programs, such as Seg3D, with large and growing user bases.
Today's Tools - Tomorrow's Scientists: An ImageVis3D Project
 The SCI Institute holds a strong belief that providing research opportunities to undergraduate and even high school students will not only encourage them to pursue studies in the sciences, but also give them a head start in their future academic lives. Being allowed to work side by side with PhD-level scientists within a real research institute moves science from something that happens in a text book or highly structured laboratory to the dynamic work environment shared by scientists around the world.
The SCI Institute holds a strong belief that providing research opportunities to undergraduate and even high school students will not only encourage them to pursue studies in the sciences, but also give them a head start in their future academic lives. Being allowed to work side by side with PhD-level scientists within a real research institute moves science from something that happens in a text book or highly structured laboratory to the dynamic work environment shared by scientists around the world.In this year's high school summer intern program, the SCI Institute invited four students, one each from Juan Diego Catholic High School, The Waterford School, and two from West High School. These students were given the opportunity to work with a lead software developer from the National Institutes of Health (NIH) sponsored Center for Integrative Biomedical Computing (CIBC). Their task seemed simple: take Seg3D and ImageVis3D (two advanced software tools developed by the CIBC), find a dataset of interest to the student, load that data, and experiment with the software on both desktop and iPad versions. And then, present your results to your high school peers. In the end, the students learned that research is a full-contact sport, not just a homework assignment. They had to 'dig-in', expand their knowledge, and learn about their subjects of interest, their data, their software, even their computers. In the end, the students translated this process and knowledge to science classes at their school. And, the top question after the presentations? Oddly enough, "how do I get an internship like yours?" Kids excited about a science internship! Mission Accomplished.
Solving Mysteries of Autism via The Power of Collaboration
Dr. Guido Gerig Early-Brain Development Research Reveals Vibrant Clues
By Peta Owens-Liston
 |
| Dr. Guido Gerig |
Pencil in hand, Gerig fills three pages with a whirl of sketches as he explains how his imaging work illuminates clinical findings in his research involving early brain development, and more specifically autism. The sketches fade to stick figure-status as Gerig jumps back and forth between the paper and the color-exploding images on his computer screen. Vivid and seemingly pulsating with life, the brain-development images are a result of thousands of highly precise, quantifiable measurements never before captured visually.
Mobile Mayhem: Researchers Harness Kraken to Model Explosions via Transport
by Gregory Scott Jones - NICS
 |
| The crater resulting from the Spanish Fork Detonation. |
Now, the good news: America's track record in transporting these materials is about as safe as they come. Very rarely, almost never in fact, are the potential dangers of these transports realized, largely due to instituted safeguards that seem to work very well.
However, accidents can happen. Take the August 2005 incident in Spanish Fork Canyon, Utah, for instance. A truck carrying 35,500 pounds of explosives—specifically small boosters used in seismic testing—overturned and exploded, creating a crater in the highway estimated to be between 20 to 35 feet deep and 70 feet wide according to the Utah Department of Transportation. But the damage wasn't solely financial. Four people, including the truck driver and a passenger, were hospitalized.
Meshing for Multimaterial Biological Volumes: BioMesh3D
With the widespread use of medical imaging, there is a growing need for better analysis of datasets. One method for improving analysis is to simulate biological processes and medical interventions in silico, in order to render better predictions. For example, the CIBC center is currently collaborating with Dr. Triedman at Children's Hospital in Boston to develop a computer model that will help guide the implantation of Implantable Cardiac Defibrillators (ICDs). This model uses pediatric imaging to select placement of electrode leads to generate the optimal field for defibrillation. One of the critical pieces in the development of the model is the generation of quality meshes for electric field simulation. Because the project is entering the validation phase where many cases need to be reviewed, a robust and automated Meshing Pipeline is required.
Atrial Fibrillation
Uncertainty Visualization
 |
| An Isosurface visualization of a magnetic resonance imaging data set (in orange) surrounded by a volume rendered region of low opacity (in green) to indicate uncertainty in surface position. |
Imaging Meets Electrophysiology
In atrial fibrillation, the upper two chambers (the left and right atria) of the heart lose their synchronization and beat erratically and inefficiently. The same condition in the lower chambers (ventricles) of the heart is fatal within minutes and defibrillators are necessary to restore coordination. In the atria, death is by stealth and occurs over years, which is both good news and bad.
 Because it is not immediately fatal, there is time to treat atrial fibrillation–but also time to ignore it. While it is not immediately life-threatening, AF does immediately reduce the pumping capacity of the heart and elevates the heart rate of the entire organ. Patients cannot be as physically active as they often wish but many adjust to the symptoms and live with the disease untreated for many years.
Because it is not immediately fatal, there is time to treat atrial fibrillation–but also time to ignore it. While it is not immediately life-threatening, AF does immediately reduce the pumping capacity of the heart and elevates the heart rate of the entire organ. Patients cannot be as physically active as they often wish but many adjust to the symptoms and live with the disease untreated for many years.
Visualization
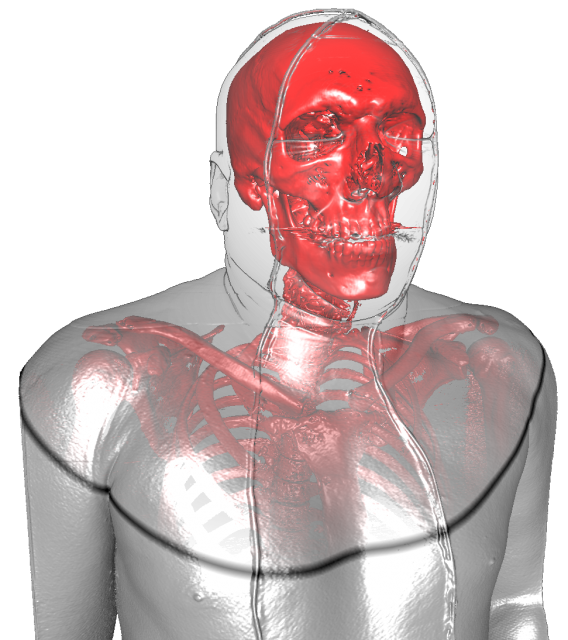 |
| Figure from T. Fogal and J. Krüger, a Clearview rendering of the visible human male dataset |
Simulation of Electric Stimulation for Bone Growth
Subject Specific, Multiscale Simulation of Electrophysiology
|
A "typical" workflow that applies to many problems in biomedical simulation contains the following elements: (i) Image acquisition and processing for a tissue, organ or region of interest (imaging and image processing), (ii) Identification of structures, tissues, cells or organelles within the images(image processing and segmentation), (iii) Fitting of geometric surfaces to the boundaries between structures and regions (geometric modelling), (iv) Generation of three-dimensional volume mesh from hexahedra or tetrahedra (meshing), and (v) Application of tissue parameters and boundary conditions and computation of spatial distribution of scalar, vector or tensor quantities of interest (simulation). |
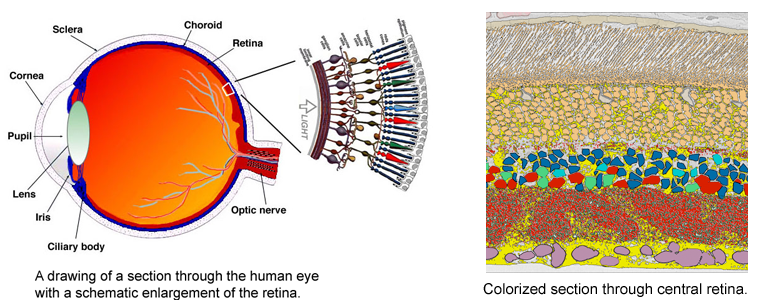
CRCNS: Fighting Blindness
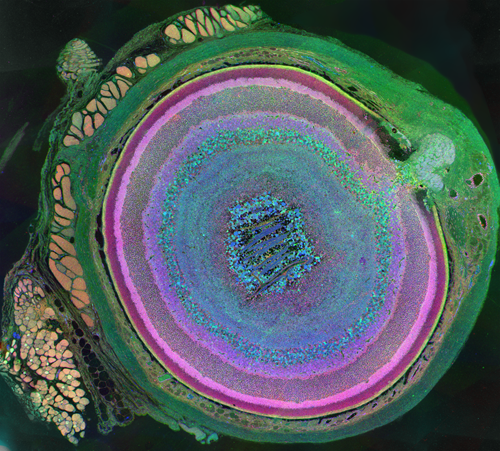 Despite great advances in neuroscience and medical technology in recent decades, nearly ten million Americans still suffer blindness due to retinal degenerative diseases such as retinitis pigmentosa (RP), age-related macular degeneration (AMD), diabetic retinopathy, and glaucoma. Unfortunately, current treatments available for these conditions are still quite limited. A primary challenge to developing effective treatments is the need for a complete understanding of the highly complex and delicate systems that compose the retina and how those systems change in response to degenerative disorders.
Despite great advances in neuroscience and medical technology in recent decades, nearly ten million Americans still suffer blindness due to retinal degenerative diseases such as retinitis pigmentosa (RP), age-related macular degeneration (AMD), diabetic retinopathy, and glaucoma. Unfortunately, current treatments available for these conditions are still quite limited. A primary challenge to developing effective treatments is the need for a complete understanding of the highly complex and delicate systems that compose the retina and how those systems change in response to degenerative disorders.Remodeling processes that occur in the neuronal pathways within the retina during the course of retinal deterioration are of particular importance to the development of treatments for these conditions. Researchers at the Robert E. Marc Laboratory at the Moran Eye Center are collaborating with the SCI Institute on a project supported by the NIH-NIBIB (grant number 5R01EB005832) to develop high-throughput techniques for reconstructing and visualizing the neural structures that compose the retina in order to meet these challenges.